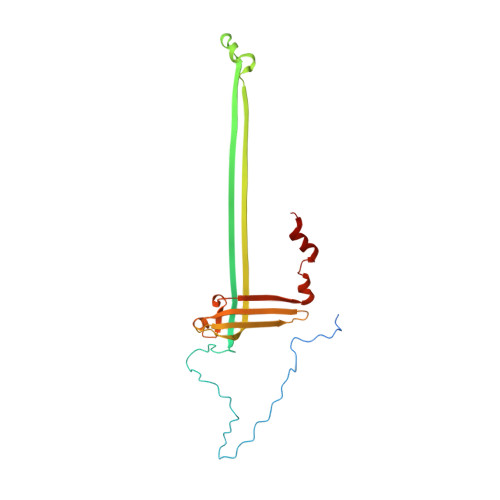
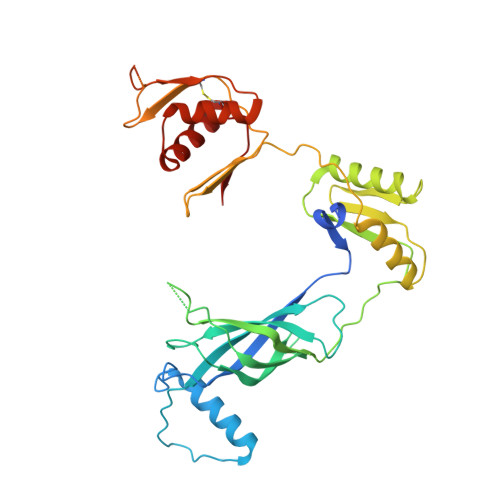
Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Tan, J., Zhang, X., Wang, X., Xu, C., Chang, S., Wu, H., Wang, T., Liang, H., Gao, H., Zhou, Y., Zhu, Y.(2021) Cell 184: 2665-2679.e19
- PubMed: 33882274
- DOI: https://doi.org/10.1016/j.cell.2021.03.057
- Primary Citation of Related Structures:
7CBL, 7CBM, 7CG0, 7CG4, 7CG7, 7CGB, 7CGO, 7E80, 7E81, 7E82 - PubMed Abstract:
The bacterial flagellar motor is a supramolecular protein machine that drives rotation of the flagellum for motility, which is essential for bacterial survival in different environments and a key determinant of pathogenicity. The detailed structure of the flagellar motor remains unknown. Here we present an atomic-resolution cryoelectron microscopy (cryo-EM) structure of the bacterial flagellar motor complexed with the hook, consisting of 175 subunits with a molecular mass of approximately 6.3 MDa. The structure reveals that 10 peptides protruding from the MS ring with the FlgB and FliE subunits mediate torque transmission from the MS ring to the rod and overcome the symmetry mismatch between the rotational and helical structures in the motor. The LP ring contacts the distal rod and applies electrostatic forces to support its rotation and torque transmission to the hook. This work provides detailed molecular insights into the structure, assembly, and torque transmission mechanisms of the flagellar motor.
- Department of Biophysics and Department of Pathology of Sir Run Run Shaw Hospital, Life Sciences Institute and School of Medicine, Zhejiang University, Hangzhou, Zhejiang 310058, China; The MOE Key Laboratory for Biosystems Homeostasis & Protection and Zhejiang Provincial Key Laboratory of Cancer Molecular Cell Biology, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang 310058, China; Institute of Microbiology, Zhejiang University, Hangzhou, Zhejiang 310058, China.
Organizational Affiliation: