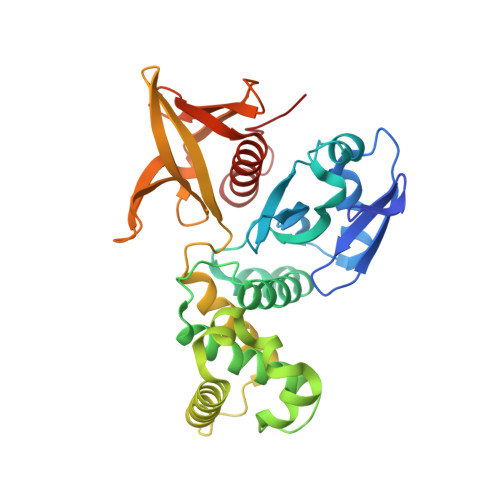
Structure of the FERM domain of a neural scaffold protein FRMPD4 implicated in X-linked intellectual disability.
Wang, M., Lin, L., Shi, Y., He, L., Wang, C., Zhu, J.(2020) Biochem J 477: 4623-4634
- PubMed: 33216857
- DOI: https://doi.org/10.1042/BCJ20200857
- Primary Citation of Related Structures:
7BYJ - PubMed Abstract:
Scaffold proteins play crucial roles in orchestrating synaptic signaling and plasticity in the excitatory synapses by providing a structural link between glutamatergic receptors, signaling molecules, and neuronal cytoskeletons. FRMPD4 is a neural scaffold protein that binds to metabotropic glutamate receptors via its FERM domain. Here, we determine the crystal structure of the FERM domain of FRMPD4 at 2.49 Å resolution. The structure reveals that the canonical target binding groove of FRMPD4 FERM is occupied by a conserved fragment C-terminal to the FERM domain, suggesting that the FRMPD4-mGluR interaction may adopt a distinct binding mode. In addition, FRMPD4 FERM does not contain a typical phosphoinositide binding site at the F1/F3 cleft found in ERM family FERM domains, but it possesses a conserved basic residue cluster on the F2 lobe which could bind to lipid effectively. Finally, analysis of mutations that are associated with X-linked intellectual disability suggests that they may compromise the biological function of FRMPD4 by destabilizing the FERM structure.
- MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027, China.
Organizational Affiliation: