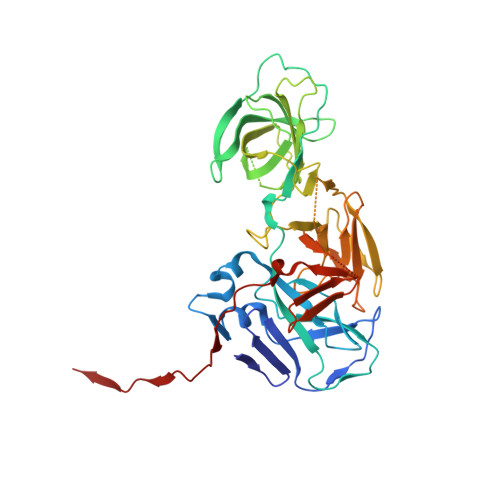
Molecular mechanism underlying selective inhibition of mRNA nuclear export by herpesvirus protein ORF10.
Feng, H., Tian, H., Wang, Y., Zhang, Q., Lin, N., Liu, S., Yu, Y., Deng, H., Gao, P.(2020) Proc Natl Acad Sci U S A 117: 26719-26727
- PubMed: 33033226
- DOI: https://doi.org/10.1073/pnas.2007774117
- Primary Citation of Related Structures:
7BYF - PubMed Abstract:
Viruses employ multiple strategies to inhibit host mRNA nuclear export. Distinct to the generally nonselective inhibition mechanisms, ORF10 from gammaherpesviruses inhibits mRNA export in a transcript-selective manner by interacting with Rae1 (RNA export 1) and Nup98 (nucleoporin 98). We now report the structure of ORF10 from MHV-68 (murine gammaherpesvirus 68) bound to the Rae1-Nup98 heterodimer, thereby revealing detailed intermolecular interactions. Structural and functional assays highlight that two highly conserved residues of ORF10, L60 and M413, play critical roles in both complex assembly and mRNA export inhibition. Interestingly, although ORF10 occupies the RNA-binding groove of Rae1-Nup98, the ORF10-Rae1-Nup98 ternary complex still maintains a comparable RNA-binding ability due to the ORF10-RNA direct interaction. Moreover, mutations on the RNA-binding surface of ORF10 disrupt its function of mRNA export inhibition. Our work demonstrates the molecular mechanism of ORF10-mediated selective inhibition and provides insights into the functions of Rae1-Nup98 in regulating host mRNA export.
- CAS Key Laboratory of Infection and Immunity, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: