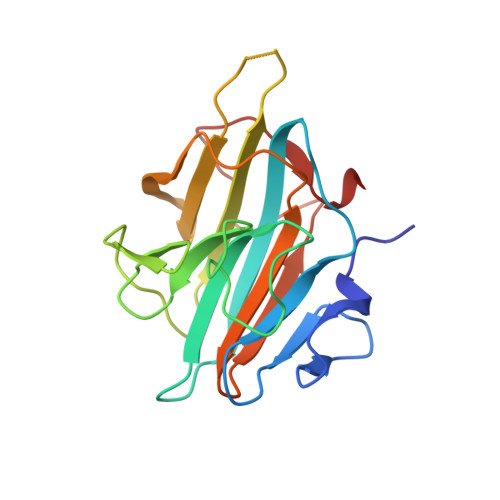
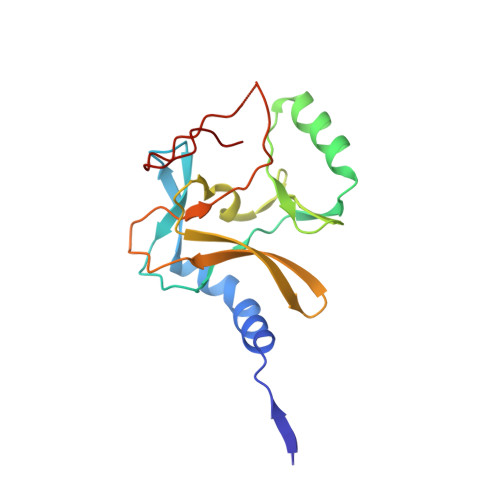
Crystal Structure of MLL2 Complex Guides the Identification of a Methylation Site on P53 Catalyzed by KMT2 Family Methyltransferases.
Li, Y., Zhao, L., Tian, X., Peng, C., Gong, F., Chen, Y.(2020) Structure 28: 1141
- PubMed: 32697937
- DOI: https://doi.org/10.1016/j.str.2020.07.002
- Primary Citation of Related Structures:
7BRE - PubMed Abstract:
KMT2 family methyltransferases methylate histone H3 lysine 4 and play essential roles in multiple cellular processes. MLL2 (KMT2B) is required for early epigenetic decisions during development and contributes to the methylation of bivalent promoters. Here, we determined the crystal structure of the MLL2 SET -RBBP5 AS-ABM -ASH2L SPRY complex and confirmed that RBBP5 AS-ABM -ASH2L SPRY was essential for activating the MLL2 SET domain through a conserved mechanism across KMT2 family complexes. In the MLL2 complex structure, a short N-terminal loop of MLL2 SET adopts a similar configuration of the H3 peptide and inserts into the substrate-binding pocket of another MLL2 SET , indicating a potential substrate for MLL2 SET . We identify that P53 contains a sequence similar to the N-terminal loop of MLL2 SET , and demonstrate that K305 of P53 could be methylated by KMT2 family complexes except for SET1A. Our results provide an important implication of functional interplay between P53 and KMT2 family complexes, and also suggest the possible broad landscape of non-histone substrate for KMT2 family methyltransferases.
- State Key Laboratory of Molecular Biology, National Center for Protein Science Shanghai, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: