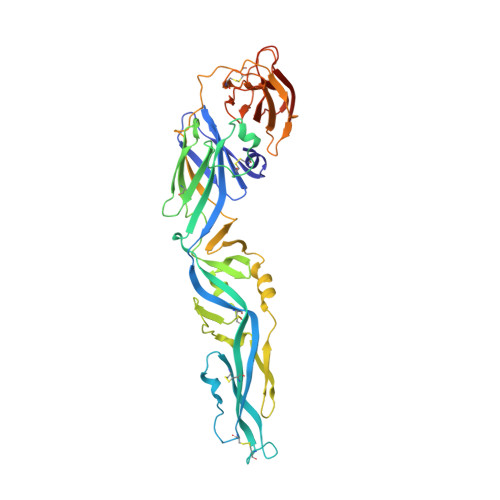
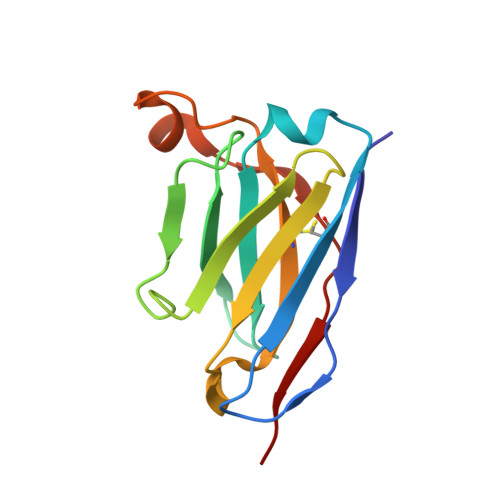
Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch.
Dai, L., Xu, K., Li, J., Huang, Q., Song, J., Han, Y., Zheng, T., Gao, P., Lu, X., Yang, H., Liu, K., Xia, Q., Wang, Q., Chai, Y., Qi, J., Yan, J., Gao, G.F.(2021) Nat Immunol 22: 958-968
- PubMed: 34267374
- DOI: https://doi.org/10.1038/s41590-021-00966-6
- Primary Citation of Related Structures:
7BPK, 7BQ5 - PubMed Abstract:
Antibody-dependent enhancement (ADE) is an important safety concern for vaccine development against dengue virus (DENV) and its antigenically related Zika virus (ZIKV) because vaccine may prime deleterious antibodies to enhance natural infections. Cross-reactive antibodies targeting the conserved fusion loop epitope (FLE) are known as the main sources of ADE. We design ZIKV immunogens engineered to change the FLE conformation but preserve neutralizing epitopes. Single vaccination conferred sterilizing immunity against ZIKV without ADE of DENV-serotype 1-4 infections and abrogated maternal-neonatal transmission in mice. Unlike the wild-type-based vaccine inducing predominately cross-reactive ADE-prone antibodies, B cell profiling revealed that the engineered vaccines switched immunodominance to dispersed patterns without DENV enhancement. The crystal structure of the engineered immunogen showed the dimeric conformation of the envelope protein with FLE disruption. We provide vaccine candidates that will prevent both ZIKV infection and infection-/vaccination-induced DENV ADE.
- CAS Key Laboratory of Pathogen Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences (CAS), Beijing, China. dailp@im.ac.cn.
Organizational Affiliation: