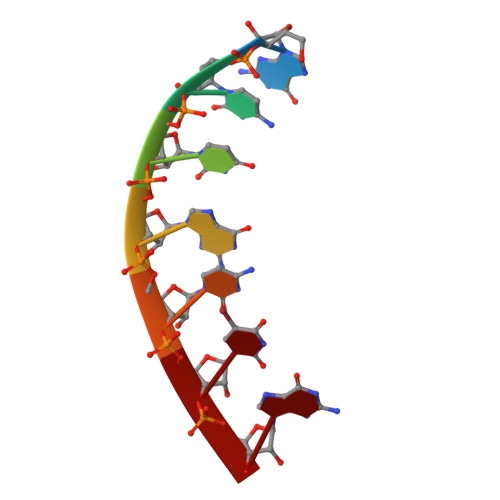
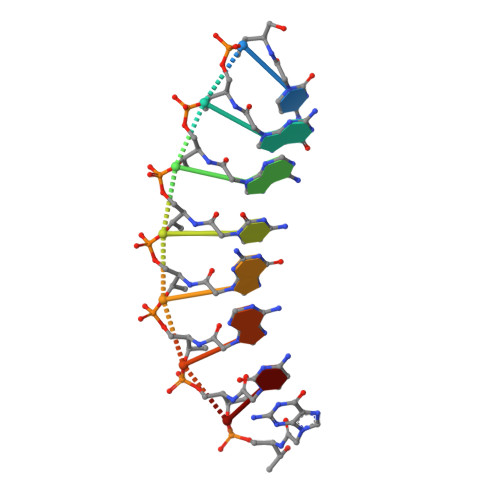
Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Kamiya, Y., Satoh, T., Kodama, A., Suzuki, T., Murayama, K., Kashida, H., Uchiyama, S., Kato, K., Asanuma, H.(2020) Commun Chem