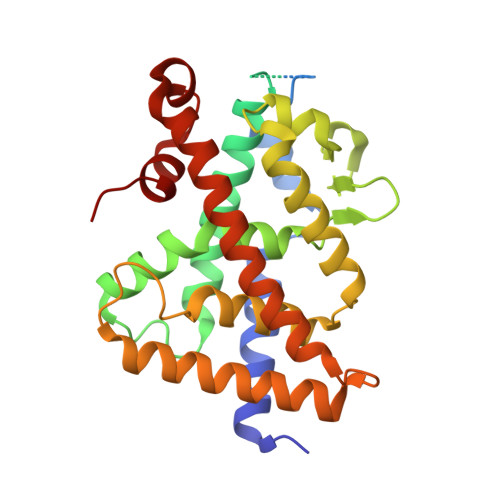
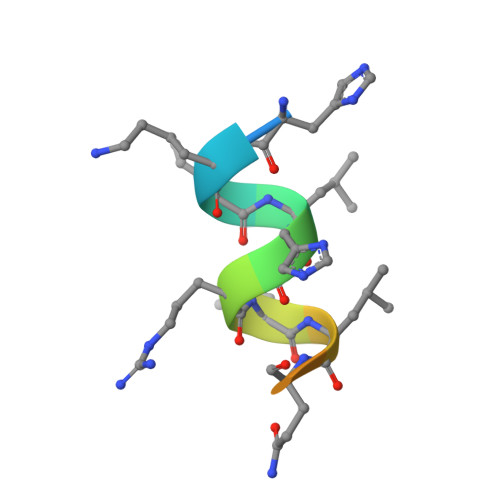
Vitamin D Analogs Bearing C-20 Modifications Stabilize the Agonistic Conformation of Non-Responsive Vitamin D Receptor Variants.
Belorusova, A.Y., Rovito, D., Chebaro, Y., Doms, S., Verlinden, L., Verstuyf, A., Metzger, D., Rochel, N., Laverny, G.(2022) Int J Mol Sci 23
- PubMed: 35955580
- DOI: https://doi.org/10.3390/ijms23158445
- Primary Citation of Related Structures:
7BNS, 7BNU - PubMed Abstract:
The Vitamin D receptor (VDR) plays a key role in calcium homeostasis, as well as in cell proliferation and differentiation. Among the large number of VDR ligands that have been developed, we have previously shown that BXL-62 and Gemini-72, two C-20-modified vitamin D analogs are highly potent VDR agonists. In this study, we show that both VDR ligands restore the transcriptional activities of VDR variants unresponsive to the natural ligand and identified in patients with rickets. The elucidated mechanisms of action underlying the activities of these C-20-modified analogs emphasize the mutual adaptation of the ligand and the VDR ligand-binding pocket.
- Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), F-67400 Illkirch, France.
Organizational Affiliation: