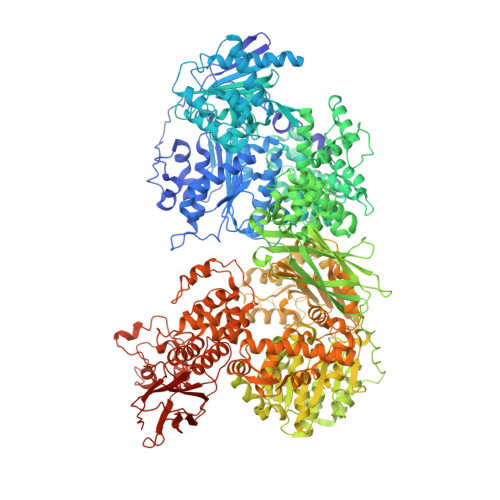
Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
Absmeier, E., Vester, K., Ghane, T., Burakovskiy, D., Milon, P., Imhof, P., Rodnina, M.V., Santos, K.F., Wahl, M.C.(2021) J Biological Chem 297: 100829-100829
- PubMed: 34048711
- DOI: https://doi.org/10.1016/j.jbc.2021.100829
- Primary Citation of Related Structures:
7BDI, 7BDJ, 7BDK, 7BDL - PubMed Abstract:
Brr2 is an essential Ski2-like RNA helicase that exhibits a unique structure among the spliceosomal helicases. Brr2 harbors a catalytically active N-terminal helicase cassette and a structurally similar but enzymatically inactive C-terminal helicase cassette connected by a linker region. Both cassettes contain a nucleotide-binding pocket, but it is unclear whether nucleotide binding in these two pockets is related. Here we use biophysical and computational methods to delineate the functional connectivity between the cassettes and determine whether occupancy of one nucleotide-binding site may influence nucleotide binding at the other cassette. Our results show that Brr2 exhibits high specificity for adenine nucleotides, with both cassettes binding ADP tighter than ATP. Adenine nucleotide affinity for the inactive C-terminal cassette is more than two orders of magnitude higher than that of the active N-terminal cassette, as determined by slow nucleotide release. Mutations at the intercassette surfaces and in the connecting linker diminish the affinity of adenine nucleotides for both cassettes. Moreover, we found that abrogation of nucleotide binding at the C-terminal cassette reduces nucleotide binding at the N-terminal cassette 70 Å away. Molecular dynamics simulations identified structural communication lines that likely mediate these long-range allosteric effects, predominantly across the intercassette interface. Together, our results reveal intricate networks of intramolecular interactions in the complex Brr2 RNA helicase, which fine-tune its nucleotide affinities and which could be exploited to regulate enzymatic activity during splicing.
- Structural Biochemistry, Freie Universität Berlin, Berlin, Germany.
Organizational Affiliation: