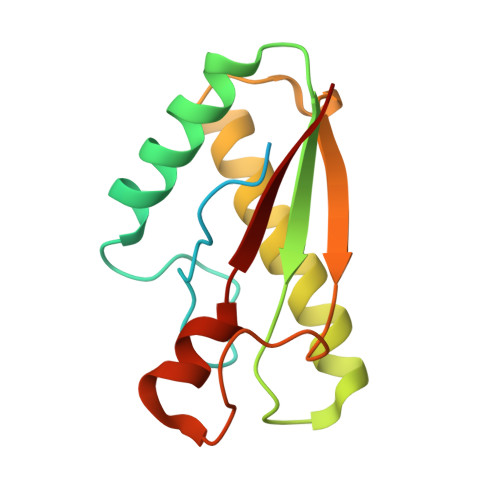
Anchoring the T6SS to the cell wall: Crystal structure of the peptidoglycan binding domain of the TagL accessory protein.
Nguyen, V.S., Spinelli, S., Cascales, E., Roussel, A., Cambillau, C., Leone, P.(2021) PLoS One 16: e0254232-e0254232
- PubMed: 34214145
- DOI: https://doi.org/10.1371/journal.pone.0254232
- Primary Citation of Related Structures:
7BBA - PubMed Abstract:
The type VI secretion system (T6SS) is a widespread mechanism of protein delivery into target cells, present in more than a quarter of all sequenced Gram-negative bacteria. The T6SS constitutes an important virulence factor, as it is responsible for targeting effectors in both prokaryotic and eukaryotic cells. The T6SS comprises a tail structure tethered to the cell envelope via a trans-envelope complex. In most T6SS, the membrane complex is anchored to the cell wall by the TagL accessory protein. In this study, we report the first crystal structure of a peptidoglycan-binding domain of TagL. The fold is conserved with members of the OmpA/Pal/MotB family, and more importantly, the peptidoglycan binding site is conserved. This structure further exemplifies how proteins involved in anchoring to the cell wall for different cellular functions rely on an interaction network with peptidoglycan strictly conserved.
- Architecture et Fonction des Macromolécules Biologiques, Aix-Marseille Université, Marseille, France.
Organizational Affiliation: