Crystal structures of phosphatidyl serine synthase PSS reveal the catalytic mechanism of CDP-DAG alcohol O-phosphatidyl transferases
Centola, M., Betz, H., Yildiz, O.(2021) Nat Commun 12: 6982
Experimental Data Snapshot
(2021) Nat Commun 12: 6982
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CDP-diacylglycerol--serine O-phosphatidyltransferase | 225 | Methanocaldococcus jannaschii DSM 2661 | Mutation(s): 0 Gene Names: pssA, MJ1212 EC: 2.7.8.8 Membrane Entity: Yes | 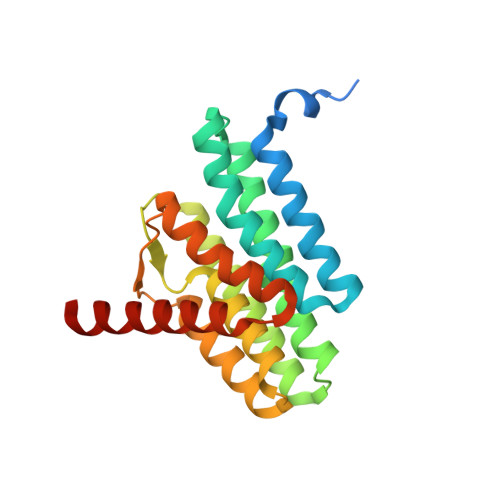 | |
UniProt | |||||
Find proteins for Q58609 (Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440)) Explore Q58609 Go to UniProtKB: Q58609 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q58609 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 58A Query on 58A | C [auth A], N [auth B] | 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine C48 H85 N3 O15 P2 WVVFFOKRFKIBHD-ZIPNUMAKSA-N |  | ||
| OLC Query on OLC | J [auth A] K [auth A] L [auth A] M [auth A] U [auth B] | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate C21 H40 O4 RZRNAYUHWVFMIP-GDCKJWNLSA-N |  | ||
| FLC Query on FLC | H [auth A], S [auth B], T [auth B] | CITRATE ANION C6 H5 O7 KRKNYBCHXYNGOX-UHFFFAOYSA-K |  | ||
| CA Query on CA | D [auth A], O [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | F [auth A], G [auth A], Q [auth B], R [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | E [auth A], P [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA Query on NA | I [auth A], W [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.314 | α = 90 |
| b = 70.758 | β = 90 |
| c = 95.545 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |