Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
Giese, C., Puorger, C., Ignatov, O., Zigova, Z., Weber, M., Scharer, M.A., Capitani, G., Glockshuber, R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chaperone protein FimC | A [auth C] | 211 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: fimC, b4316, JW4279 | 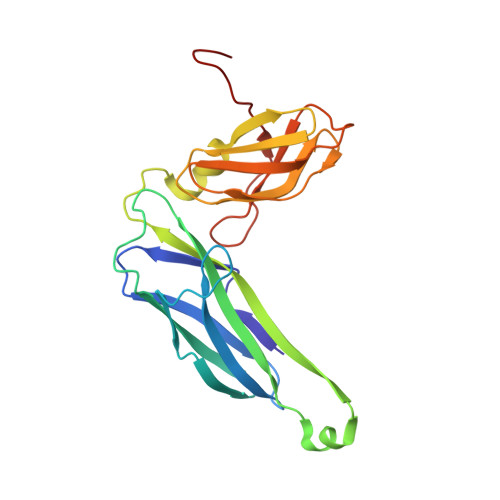 |
UniProt | |||||
Find proteins for P31697 (Escherichia coli (strain K12)) Explore P31697 Go to UniProtKB: P31697 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P31697 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fimbrin-like protein FimI | B [auth I] | 140 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: fimI, b4315, JW5779 | 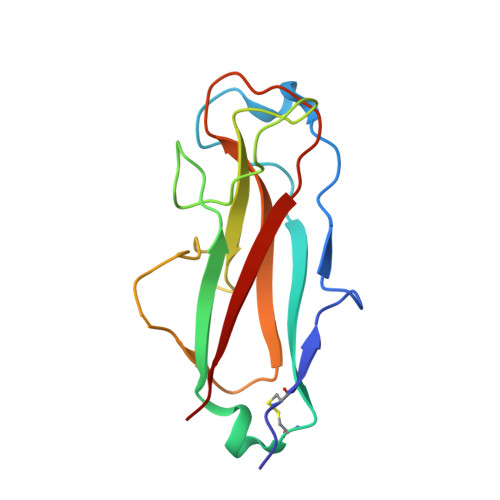 |
UniProt | |||||
Find proteins for P39264 (Escherichia coli (strain K12)) Explore P39264 Go to UniProtKB: P39264 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P39264 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Outer membrane usher protein FimD | C [auth D] | 125 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: fimD, b4317, JW5780 | 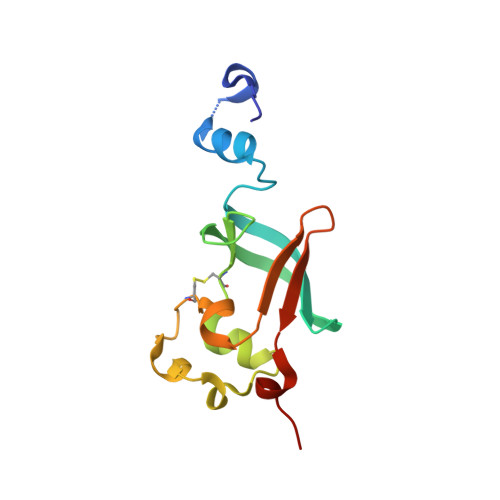 |
UniProt | |||||
Find proteins for P30130 (Escherichia coli (strain K12)) Explore P30130 Go to UniProtKB: P30130 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30130 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EDO Query on EDO | D | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 35.24 | α = 90 |
| b = 104.16 | β = 90 |
| c = 132.16 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XSCALE | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | -- |