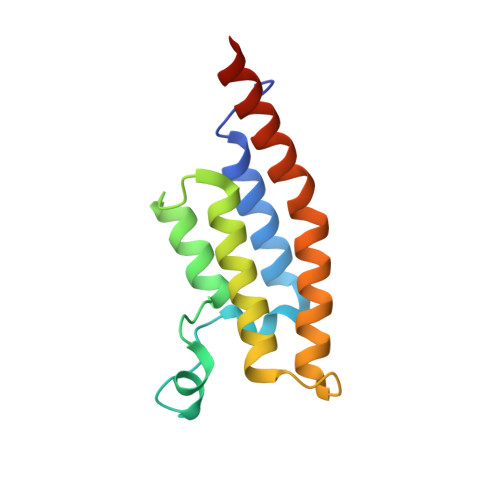
Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
Krojer, T., Talon, R., Fairhead, M., Szykowska, A., Burgess-Brown, N.A., Brennan, P.E., Arrowsmith, C.H., Edwards, A.M., Bountra, C., von Delft, F.To be published.