Crystal structure of the metallo-beta-lactamase VIM2 with 139
Brem, J., Schofield, C.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-lactamase VIM-2 | 242 | Pseudomonas aeruginosa | Mutation(s): 0 Gene Names: blaVIM-2, bla vim-2, bla-VIM-2, blasVIM-2, blaVIM2, blm, VIM-2, PAERUG_P32_London_17_VIM_2_10_11_06255 EC: 3.5.2.6 | 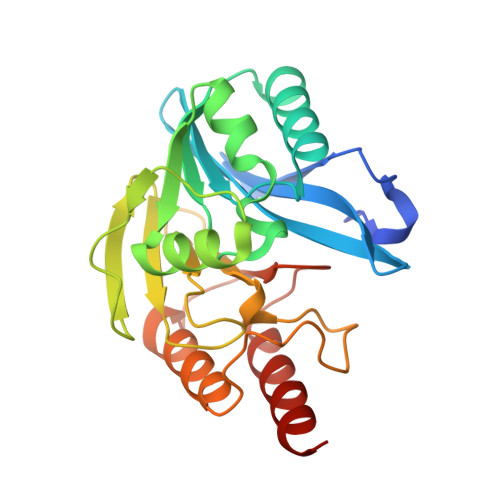 | |
UniProt | |||||
Find proteins for Q9K2N0 (Pseudomonas aeruginosa) Explore Q9K2N0 Go to UniProtKB: Q9K2N0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9K2N0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| R9K (Subject of Investigation/LOI) Query on R9K | F [auth A], M [auth B] | 3-(2-chlorophenyl)-7-methyl-1~{H}-indole-2-carboxylic acid C16 H12 Cl N O2 CXZJJQHOJSSHGW-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | C [auth A] D [auth A] E [auth A] J [auth B] K [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| FMT Query on FMT | G [auth A] H [auth A] I [auth A] N [auth B] O [auth B] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 103.251 | α = 90 |
| b = 79.066 | β = 130.37 |
| c = 67.951 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Innovative Medicines Initiative | United Kingdom | Enable |