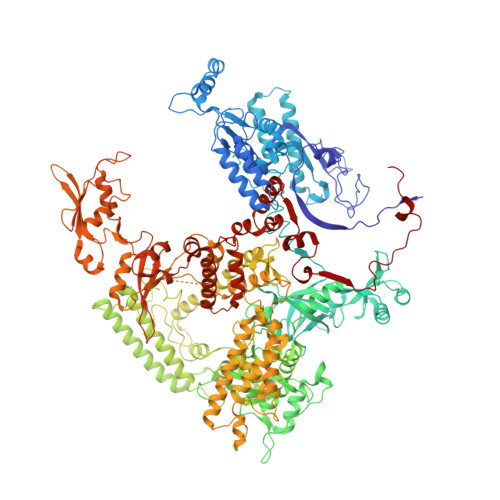
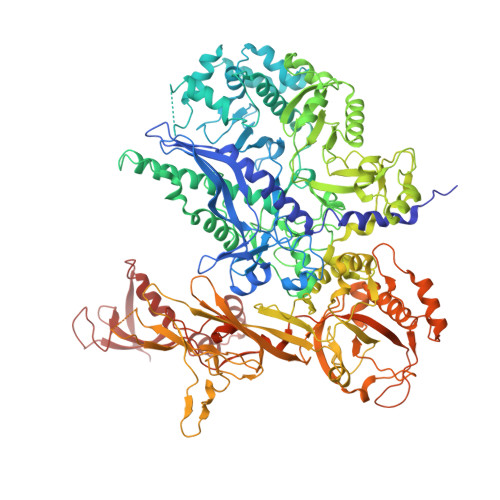
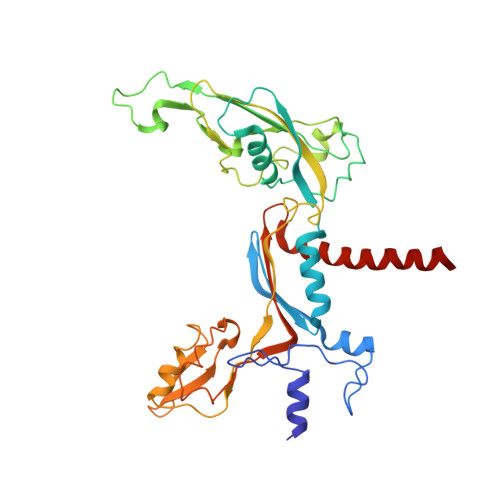
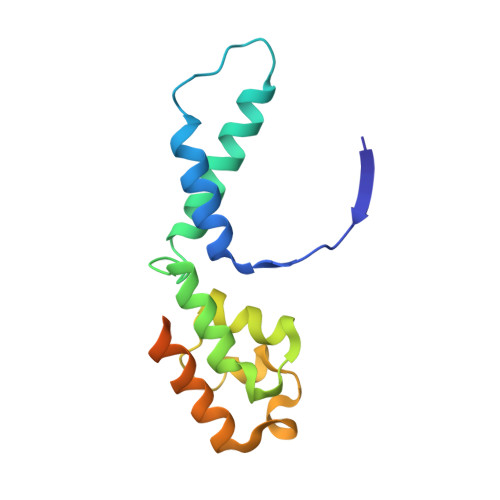
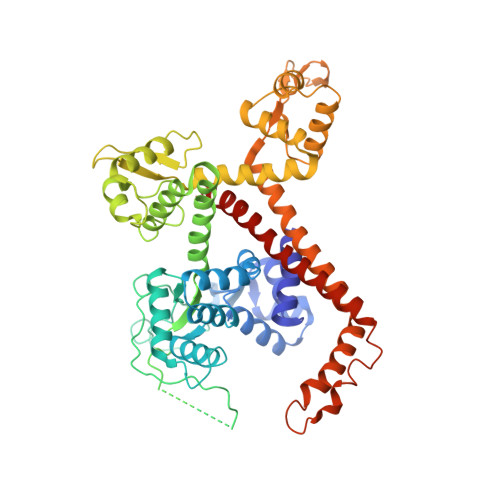
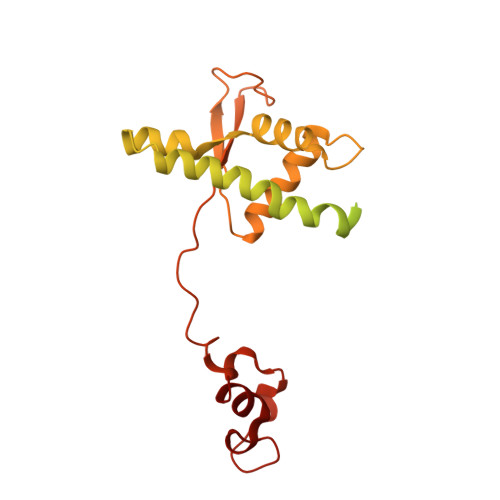
Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Girbig, M., Misiaszek, A.D., Vorlander, M.K., Lafita, A., Grotsch, H., Baudin, F., Bateman, A., Muller, C.W.(2021) Nat Struct Mol Biol 28: 210-219
- PubMed: 33558764
- DOI: https://doi.org/10.1038/s41594-020-00555-5
- Primary Citation of Related Structures:
7A6H, 7AE1, 7AE3, 7AEA - PubMed Abstract:
RNA polymerase III (Pol III) synthesizes transfer RNAs and other short, essential RNAs. Human Pol III misregulation is linked to tumor transformation, neurodegenerative and developmental disorders, and increased sensitivity to viral infections. Here, we present cryo-electron microscopy structures at 2.8 to 3.3 Å resolution of transcribing and unbound human Pol III. We observe insertion of the TFIIS-like subunit RPC10 into the polymerase funnel, providing insights into how RPC10 triggers transcription termination. Our structures resolve elements absent from Saccharomyces cerevisiae Pol III such as the winged-helix domains of RPC5 and an iron-sulfur cluster, which tethers the heterotrimer subcomplex to the core. The cancer-associated RPC7α isoform binds the polymerase clamp, potentially interfering with Pol III inhibition by tumor suppressor MAF1, which may explain why overexpressed RPC7α enhances tumor transformation. Finally, the human Pol III structure allows mapping of disease-related mutations and may contribute to the development of inhibitors that selectively target Pol III for therapeutic interventions.
- European Molecular Biology Laboratory (EMBL), Structural and Computational Biology Unit, Heidelberg, Germany.
Organizational Affiliation: