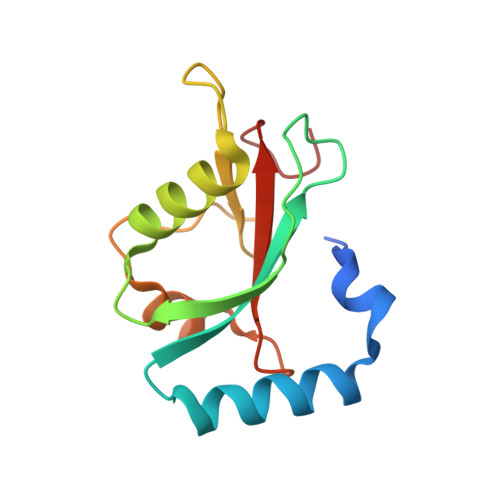
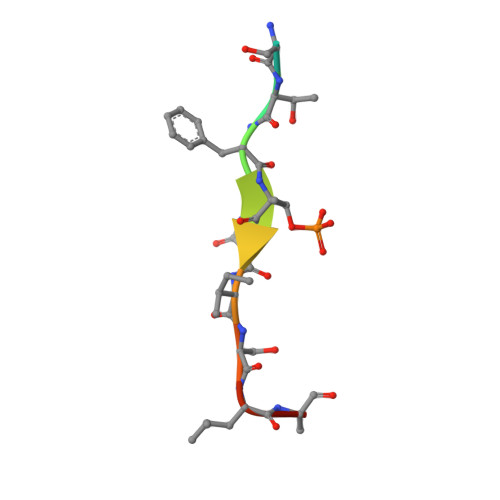
Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
Wirth, M., Mouilleron, S., Zhang, W., Sjottem, E., Princely Abudu, Y., Jain, A., Lauritz Olsvik, H., Bruun, J.A., Razi, M., Jefferies, H.B.J., Lee, R., Joshi, D., O'Reilly, N., Johansen, T., Tooze, S.A.(2021) J Mol Biology 433: 166987-166987
- PubMed: 33845085
- DOI: https://doi.org/10.1016/j.jmb.2021.166987
- Primary Citation of Related Structures:
7AA7, 7AA8, 7AA9 - PubMed Abstract:
Autophagy is a highly conserved degradative pathway, essential for cellular homeostasis and implicated in diseases including cancer and neurodegeneration. Autophagy-related 8 (ATG8) proteins play a central role in autophagosome formation and selective delivery of cytoplasmic cargo to lysosomes by recruiting autophagy adaptors and receptors. The LC3-interacting region (LIR) docking site (LDS) of ATG8 proteins binds to LIR motifs present in autophagy adaptors and receptors. LIR-ATG8 interactions can be highly selective for specific mammalian ATG8 family members (LC3A-C, GABARAP, and GABARAPL1-2) and how this specificity is generated and regulated is incompletely understood. We have identified a LIR motif in the Golgi protein SCOC (short coiled-coil protein) exhibiting strong binding to GABARAP, GABARAPL1, LC3A and LC3C. The residues within and surrounding the core LIR motif of the SCOC LIR domain were phosphorylated by autophagy-related kinases (ULK1-3, TBK1) increasing specifically LC3 family binding. More distant flanking residues also contributed to ATG8 binding. Loss of these residues was compensated by phosphorylation of serine residues immediately adjacent to the core LIR motif, indicating that the interactions of the flanking LIR regions with the LDS are important and highly dynamic. Our comprehensive structural, biophysical and biochemical analyses support and provide novel mechanistic insights into how phosphorylation of LIR domain residues regulates the affinity and binding specificity of ATG8 proteins towards autophagy adaptors and receptors.
- Molecular Cell Biology of Autophagy, The Francis Crick Institute, 1 Midland Road, London NW1 1AT, UK; Department of Cell and Chemical Biology, Leiden University Medical Center, Einthovenweg 20, 2333 ZC Leiden, the Netherlands(‡). Electronic address: m.b.wirth@lumc.nl.
Organizational Affiliation: