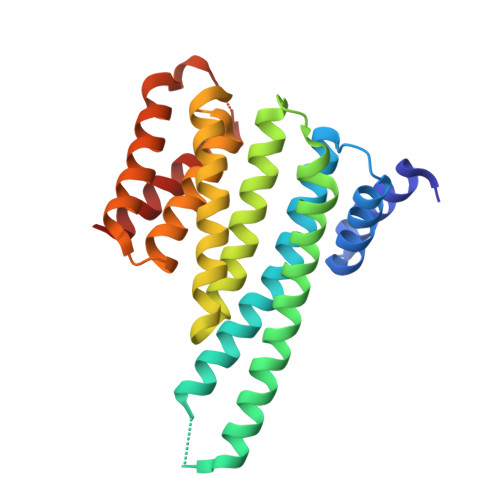
14-3-3 proteins inactivate DAPK2 by promoting its dimerization and protecting key regulatory phosphosites.
Horvath, M., Petrvalska, O., Herman, P., Obsilova, V., Obsil, T.(2021) Commun Biol 4: 986-986
- PubMed: 34413451
- DOI: https://doi.org/10.1038/s42003-021-02518-y
- Primary Citation of Related Structures:
7A6R, 7A6Y - PubMed Abstract:
Death-associated protein kinase 2 (DAPK2) is a CaM-regulated Ser/Thr protein kinase, involved in apoptosis, autophagy, granulocyte differentiation and motility regulation, whose activity is controlled by autoinhibition, autophosphorylation, dimerization and interaction with scaffolding proteins 14-3-3. However, the structural basis of 14-3-3-mediated DAPK2 regulation remains unclear. Here, we structurally and biochemically characterize the full-length human DAPK2:14-3-3 complex by combining several biophysical techniques. The results from our X-ray crystallographic analysis revealed that Thr369 phosphorylation at the DAPK2 C terminus creates a high-affinity canonical mode III 14-3-3-binding motif, further enhanced by the diterpene glycoside Fusicoccin A. Moreover, concentration-dependent DAPK2 dimerization is disrupted by Ca 2+ /CaM binding and stabilized by 14-3-3 binding in solution, thereby protecting the DAPK2 inhibitory autophosphorylation site Ser318 against dephosphorylation and preventing Ca 2+ /CaM binding. Overall, our findings provide mechanistic insights into 14-3-3-mediated DAPK2 inhibition and highlight the potential of the DAPK2:14-3-3 complex as a target for anti-inflammatory therapies.
- Department of Physical and Macromolecular Chemistry, Faculty of Science, Charles University, Prague, Czech Republic.
Organizational Affiliation: