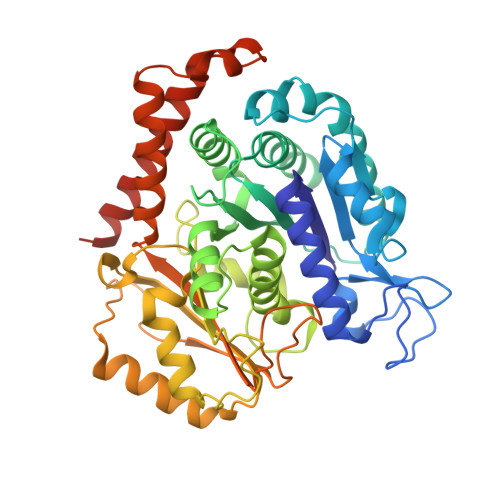
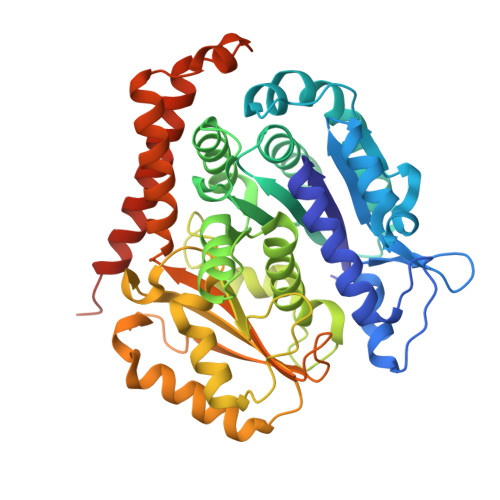
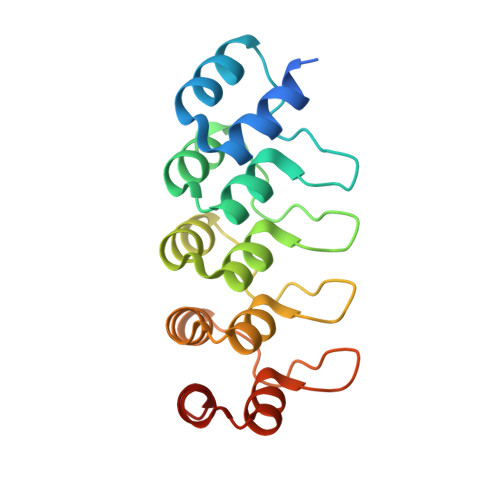
A Robust, GFP-Orthogonal Photoswitchable Inhibitor Scaffold Extends Optical Control over the Microtubule Cytoskeleton.
Gao, L., Meiring, J.C.M., Kraus, Y., Wranik, M., Weinert, T., Pritzl, S.D., Bingham, R., Ntouliou, E., Jansen, K.I., Olieric, N., Standfuss, J., Kapitein, L.C., Lohmuller, T., Ahlfeld, J., Akhmanova, A., Steinmetz, M.O., Thorn-Seshold, O.(2021) Cell Chem Biol 28: 228
- PubMed: 33275880
- DOI: https://doi.org/10.1016/j.chembiol.2020.11.007
- Primary Citation of Related Structures:
6ZWB, 6ZWC - PubMed Abstract:
Optically controlled chemical reagents, termed "photopharmaceuticals," are powerful tools for precise spatiotemporal control of proteins particularly when genetic methods, such as knockouts or optogenetics are not viable options. However, current photopharmaceutical scaffolds, such as azobenzenes are intolerant of GFP/YFP imaging and are metabolically labile, posing severe limitations for biological use. We rationally designed a photoswitchable "SBT" scaffold to overcome these problems, then derivatized it to create exceptionally metabolically robust and fully GFP/YFP-orthogonal "SBTub" photopharmaceutical tubulin inhibitors. Lead compound SBTub3 allows temporally reversible, cell-precise, and even subcellularly precise photomodulation of microtubule dynamics, organization, and microtubule-dependent processes. By overcoming the previous limitations of microtubule photopharmaceuticals, SBTubs offer powerful applications in cell biology, and their robustness and druglikeness are favorable for intracellular biological control in in vivo applications. We furthermore expect that the robustness and imaging orthogonality of the SBT scaffold will inspire other derivatizations directed at extending the photocontrol of a range of other biological targets.
- Department of Pharmacy, Ludwig-Maximilians University of Munich, Munich 81377, Germany.
Organizational Affiliation: