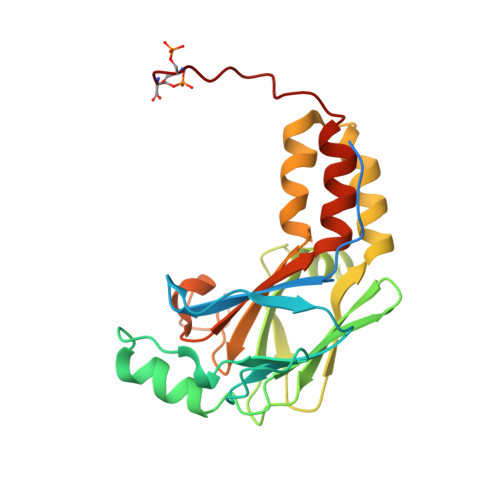
Mutations in SKI in Shprintzen-Goldberg syndrome lead to attenuated TGF-beta responses through SKI stabilization.
Gori, I., George, R., Purkiss, A.G., Strohbuecker, S., Randall, R.A., Ogrodowicz, R., Carmignac, V., Faivre, L., Joshi, D., Kjaer, S., Hill, C.S.(2021) Elife 10
- PubMed: 33416497
- DOI: https://doi.org/10.7554/eLife.63545
- Primary Citation of Related Structures:
6ZVQ - PubMed Abstract:
Shprintzen-Goldberg syndrome (SGS) is a multisystemic connective tissue disorder, with considerable clinical overlap with Marfan and Loeys-Dietz syndromes. These syndromes have commonly been associated with enhanced TGF-β signaling. In SGS patients, heterozygous point mutations have been mapped to the transcriptional co-repressor SKI, which is a negative regulator of TGF-β signaling that is rapidly degraded upon ligand stimulation. The molecular consequences of these mutations, however, are not understood. Here we use a combination of structural biology, genome editing, and biochemistry to show that SGS mutations in SKI abolish its binding to phosphorylated SMAD2 and SMAD3. This results in stabilization of SKI and consequently attenuation of TGF-β responses, both in knockin cells expressing an SGS mutation and in fibroblasts from SGS patients. Thus, we reveal that SGS is associated with an attenuation of TGF-β-induced transcriptional responses, and not enhancement, which has important implications for other Marfan-related syndromes.
- Developmental Signalling Laboratory, The Francis Crick Institute, London, United Kingdom.
Organizational Affiliation: