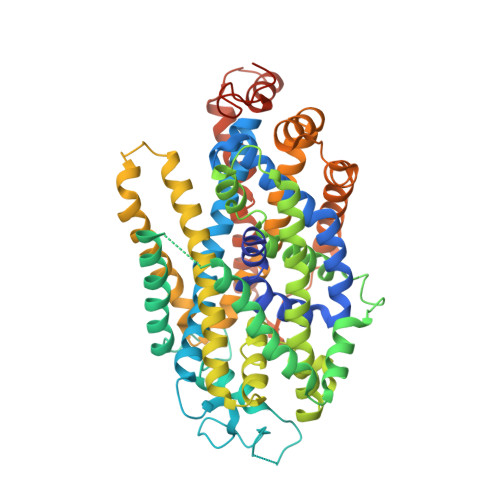
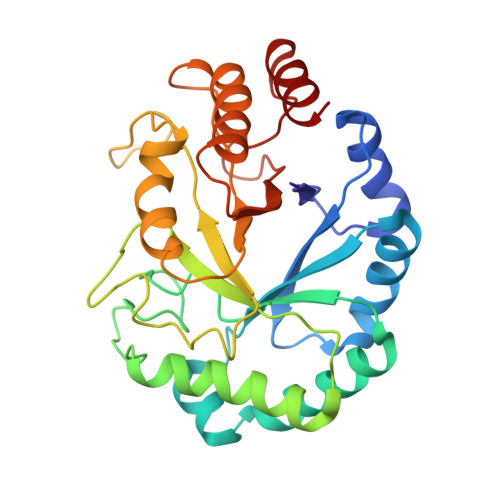
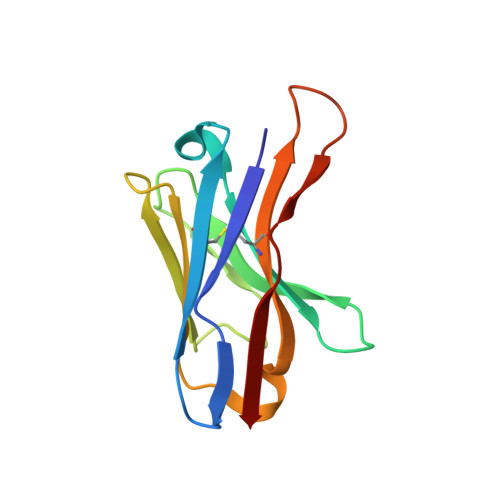
Structural insights into the inhibition of glycine reuptake.
Shahsavar, A., Stohler, P., Bourenkov, G., Zimmermann, I., Siegrist, M., Guba, W., Pinard, E., Sinning, S., Seeger, M.A., Schneider, T.R., Dawson, R.J.P., Nissen, P.(2021) Nature 591: 677-681
- PubMed: 33658720
- DOI: https://doi.org/10.1038/s41586-021-03274-z
- Primary Citation of Related Structures:
6ZBV, 6ZPL - PubMed Abstract:
The human glycine transporter 1 (GlyT1) regulates glycine-mediated neuronal excitation and inhibition through the sodium- and chloride-dependent reuptake of glycine 1-3 . Inhibition of GlyT1 prolongs neurotransmitter signalling, and has long been a key strategy in the development of therapies for a broad range of disorders of the central nervous system, including schizophrenia and cognitive impairments 4 . Here, using a synthetic single-domain antibody (sybody) and serial synchrotron crystallography, we have determined the structure of GlyT1 in complex with a benzoylpiperazine chemotype inhibitor at 3.4 Å resolution. We find that the inhibitor locks GlyT1 in an inward-open conformation and binds at the intracellular gate of the release pathway, overlapping with the glycine-release site. The inhibitor is likely to reach GlyT1 from the cytoplasmic leaflet of the plasma membrane. Our results define the mechanism of inhibition and enable the rational design of new, clinically efficacious GlyT1 inhibitors.
- Danish Research Institute of Translational Neuroscience-DANDRITE, Nordic EMBL Partnership for Molecular Medicine, Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark.
Organizational Affiliation: