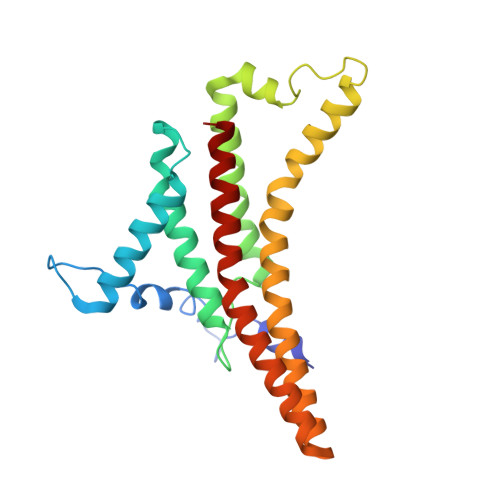
Structure of the dimeric ATP synthase from bovine mitochondria.
Spikes, T.E., Montgomery, M.G., Walker, J.E.(2020) Proc Natl Acad Sci U S A 117: 23519-23526
- PubMed: 32900941
- DOI: https://doi.org/10.1073/pnas.2013998117
- Primary Citation of Related Structures:
6YY0, 6Z1R, 6Z1U, 6ZBB, 6ZG7, 6ZG8, 6ZIK, 6ZIQ, 6ZIT, 6ZIU, 6ZMR, 6ZNA, 6ZPO, 6ZQM, 6ZQN - PubMed Abstract:
The structure of the dimeric ATP synthase from bovine mitochondria determined in three rotational states by electron cryo-microscopy provides evidence that the proton uptake from the mitochondrial matrix via the proton inlet half channel proceeds via a Grotthus mechanism, and a similar mechanism may operate in the exit half channel. The structure has given information about the architecture and mechanical constitution and properties of the peripheral stalk, part of the membrane extrinsic region of the stator, and how the action of the peripheral stalk damps the side-to-side rocking motions that occur in the enzyme complex during the catalytic cycle. It also describes wedge structures in the membrane domains of each monomer, where the skeleton of each wedge is provided by three α-helices in the membrane domains of the b-subunit to which the supernumerary subunits e, f, and g and the membrane domain of subunit A6L are bound. Protein voids in the wedge are filled by three specifically bound cardiolipin molecules and two other phospholipids. The external surfaces of the wedges link the monomeric complexes together into the dimeric structures and provide a pivot to allow the monomer-monomer interfaces to change during catalysis and to accommodate other changes not related directly to catalysis in the monomer-monomer interface that occur in mitochondrial cristae. The structure of the bovine dimer also demonstrates that the structures of dimeric ATP synthases in a tetrameric porcine enzyme have been seriously misinterpreted in the membrane domains.
- The Medical Research Council Mitochondrial Biology Unit, Cambridge Biomedical Campus, University of Cambridge, Cambridge CB2 0XY, United Kingdom.
Organizational Affiliation: