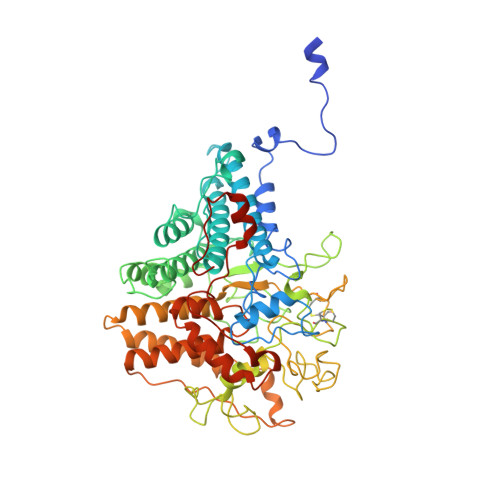
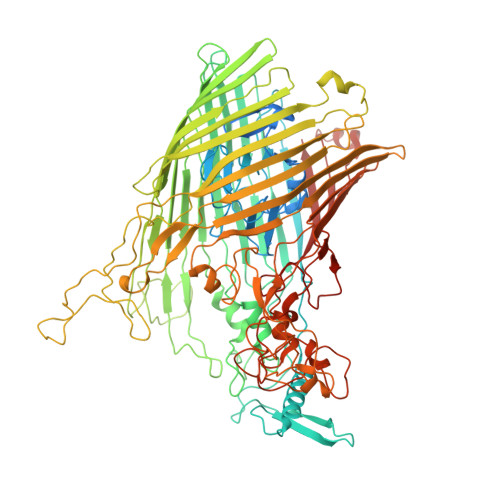
Insights into SusCD-mediated glycan import by a prominent gut symbiont.
Gray, D.A., White, J.B.R., Oluwole, A.O., Rath, P., Glenwright, A.J., Mazur, A., Zahn, M., Basle, A., Morland, C., Evans, S.L., Cartmell, A., Robinson, C.V., Hiller, S., Ranson, N.A., Bolam, D.N., van den Berg, B.(2021) Nat Commun 12: 44-44
- PubMed: 33398001
- DOI: https://doi.org/10.1038/s41467-020-20285-y
- Primary Citation of Related Structures:
6YTC, 6Z8I, 6Z9A, 6ZAZ, 6ZLT, 6ZLU, 6ZM1 - PubMed Abstract:
In Bacteroidetes, one of the dominant phyla of the mammalian gut, active uptake of large nutrients across the outer membrane is mediated by SusCD protein complexes via a "pedal bin" transport mechanism. However, many features of SusCD function in glycan uptake remain unclear, including ligand binding, the role of the SusD lid and the size limit for substrate transport. Here we characterise the β2,6 fructo-oligosaccharide (FOS) importing SusCD from Bacteroides thetaiotaomicron (Bt1762-Bt1763) to shed light on SusCD function. Co-crystal structures reveal residues involved in glycan recognition and suggest that the large binding cavity can accommodate several substrate molecules, each up to ~2.5 kDa in size, a finding supported by native mass spectrometry and isothermal titration calorimetry. Mutational studies in vivo provide functional insights into the key structural features of the SusCD apparatus and cryo-EM of the intact dimeric SusCD complex reveals several distinct states of the transporter, directly visualising the dynamics of the pedal bin transport mechanism.
- Biosciences Institute, The Medical School, Newcastle University, Newcastle upon Tyne, NE2 4HH, UK.
Organizational Affiliation: