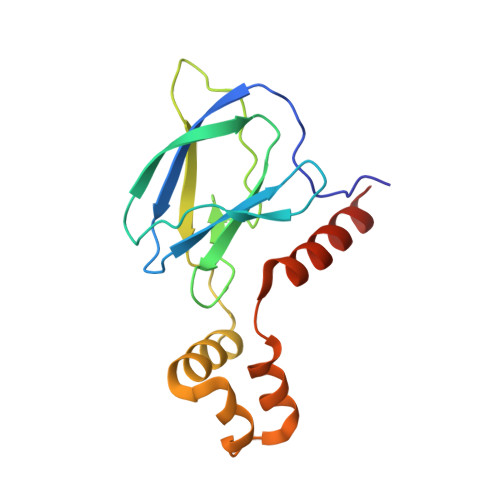
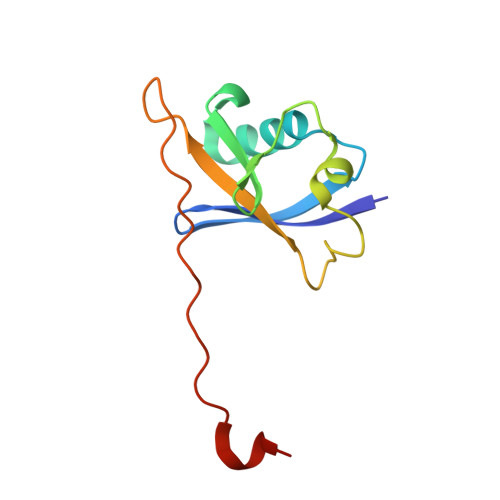
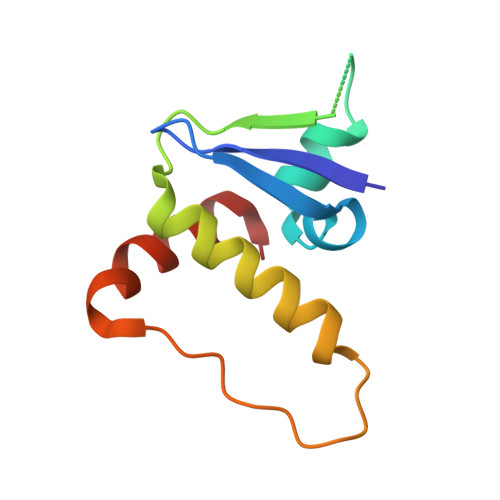
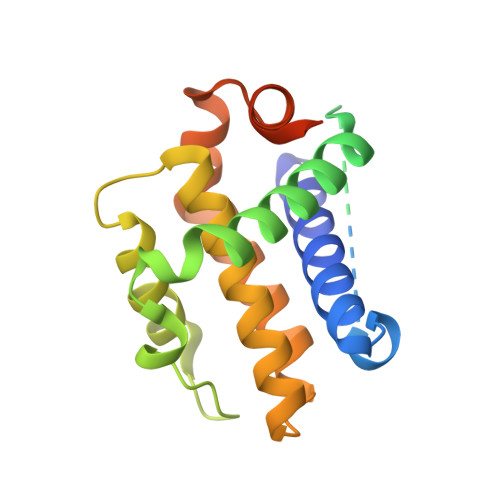
Structural Insights into PROTAC-Mediated Degradation of Bcl-xL.
Chung, C.W., Dai, H., Fernandez, E., Tinworth, C.P., Churcher, I., Cryan, J., Denyer, J., Harling, J.D., Konopacka, A., Queisser, M.A., Tame, C.J., Watt, G., Jiang, F., Qian, D., Benowitz, A.B.(2020) ACS Chem Biol 15: 2316-2323
- PubMed: 32697072
- DOI: https://doi.org/10.1021/acschembio.0c00266
- Primary Citation of Related Structures:
6ZHC - PubMed Abstract:
The Bcl-2 family of proteins, such as Bcl-xL and Bcl-2, play key roles in cancer cell survival. Structural studies of Bcl-xL formed the foundation for the development of the first Bcl-2 family inhibitors and FDA approved drugs. Recently, Pro teolysis Ta rgeting C himeras (PROTACs) that degrade Bcl-xL have been proposed as a therapeutic modality with the potential to enhance potency and reduce toxicity versus antagonists. However, no ternary complex structures of Bcl-xL with a PROTAC and an E3 ligase have been successfully determined to guide this approach. Herein, we report the design, characterization, and X-ray structure of a VHL E3 ligase-recruiting Bcl-xL PROTAC degrader. The 1.9 Å heterotetrameric structure, composed of (ElonginB:ElonginC:VHL):PROTAC:Bcl-xL, reveals an extensive network of neo-interactions, between the E3 ligase and the target protein, and between noncognate parts of the PROTAC and partner proteins. This work illustrates the challenges associated with the rational design of bifunctional molecules where interactions involve composite interfaces.
- Protein, Cellular & Structural Sciences, GlaxoSmithKline, Gunnels Wood Road, Stevenage, SG1 2NY, United Kingdom.
Organizational Affiliation: