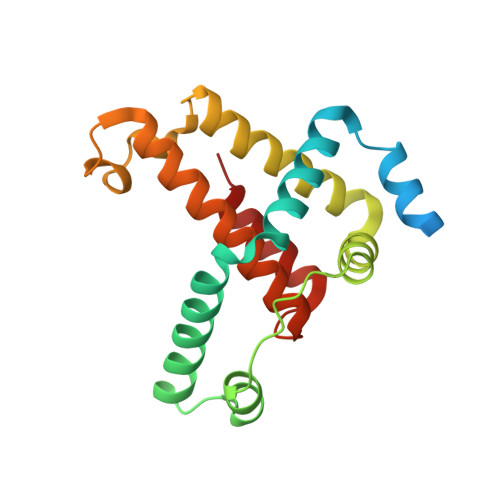
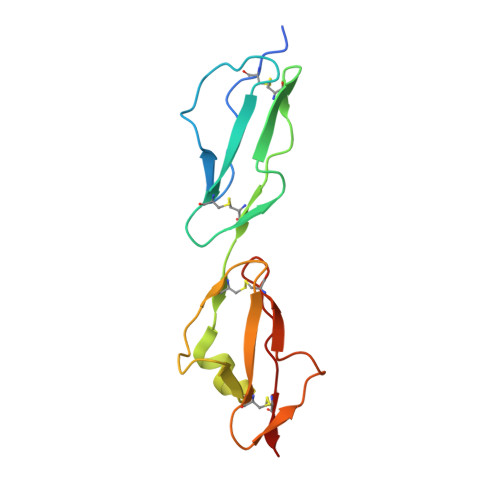
Mechanism of Borrelia immune evasion by FhbA-related proteins.
Kogan, K., Haapasalo, K., Kotila, T., Moore, R., Lappalainen, P., Goldman, A., Meri, T.(2022) PLoS Pathog 18: e1010338-e1010338
- PubMed: 35303742
- DOI: https://doi.org/10.1371/journal.ppat.1010338
- Primary Citation of Related Structures:
6ZH1 - PubMed Abstract:
Immune evasion facilitates survival of Borrelia, leading to infections like relapsing fever and Lyme disease. Important mechanism for complement evasion is acquisition of the main host complement inhibitor, factor H (FH). By determining the 2.2 Å crystal structure of Factor H binding protein A (FhbA) from Borrelia hermsii in complex with FH domains 19-20, combined with extensive mutagenesis, we identified the structural mechanism by which B. hermsii utilizes FhbA in immune evasion. Moreover, structure-guided sequence database analysis identified a new family of FhbA-related immune evasion molecules from Lyme disease and relapsing fever Borrelia. Conserved FH-binding mechanism within the FhbA-family was verified by analysis of a novel FH-binding protein from B. duttonii. By sequence analysis, we were able to group FH-binding proteins of Borrelia into four distinct phyletic types and identified novel putative FH-binding proteins. The conserved FH-binding mechanism of the FhbA-related proteins could aid in developing new approaches to inhibit virulence and complement resistance in Borrelia.
- HiLife Institute of Biotechnology, University of Helsinki, Helsinki, Finland.
Organizational Affiliation: