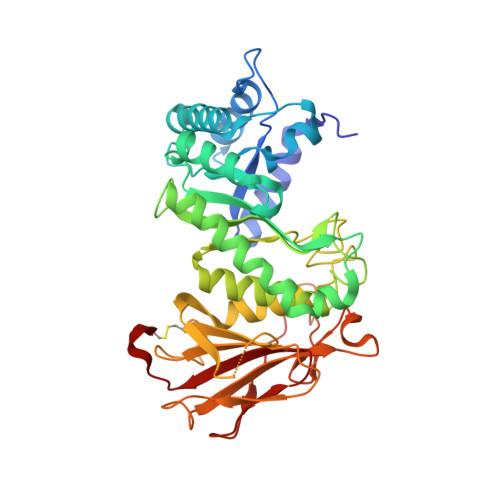
Structural insights into heparanase activity using a fluorogenic heparan sulfate disaccharide.
Wu, L., Wimmer, N., Davies, G.J., Ferro, V.(2020) Chem Commun (Camb) 56: 13780-13783
- PubMed: 33073275
- DOI: https://doi.org/10.1039/d0cc05932c
- Primary Citation of Related Structures:
6ZDM - PubMed Abstract:
A synthetic heparan sulfate disaccharide has been assessed as a fluorogenic heparanase substrate, enabling enzyme turnover and inhibition kinetics measurements despite slow turnover. Crystal structures with human heparanase also provide the first ever observation of a substrate in an activated 1S3 conformation, highlighting previously unknown interactions involved in enzymatic processing. Our data provide insights into the heparanase catalytic mechanism, and will inform the design of improved heparanase substrates and inhibitors.
- Department of Chemistry, University of York, Heslington, York, YO10 5DD, UK. gideon.davies@york.ac.uk.
Organizational Affiliation: