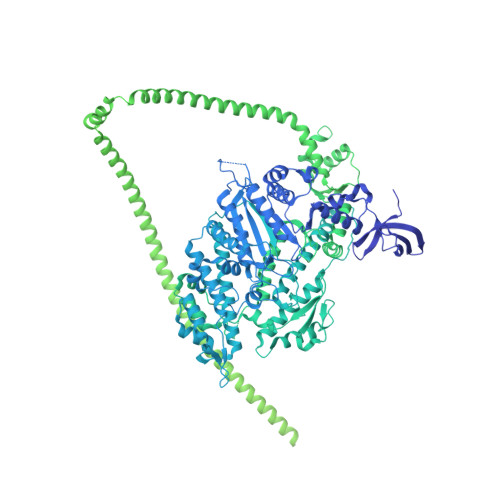
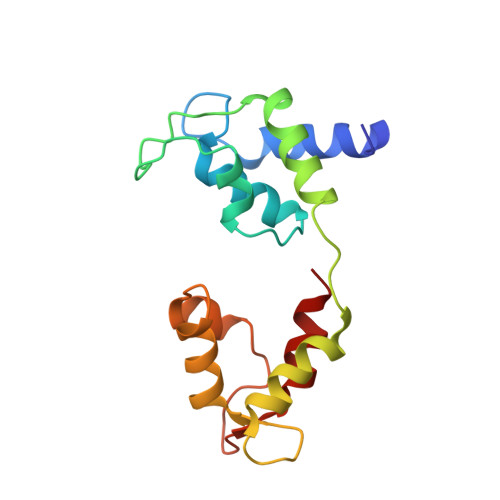
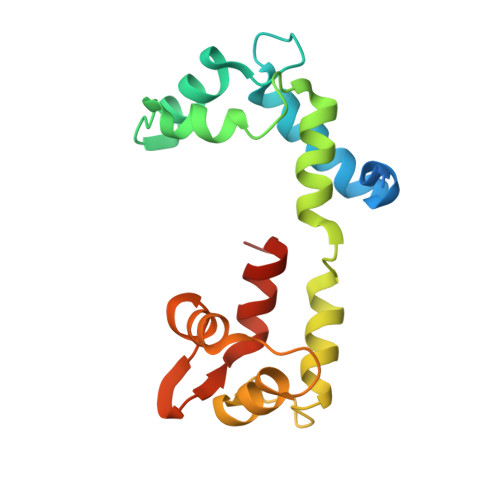
Structure of the shutdown state of myosin-2.
Scarff, C.A., Carrington, G., Casas-Mao, D., Chalovich, J.M., Knight, P.J., Ranson, N.A., Peckham, M.(2020) Nature 588: 515-520
- PubMed: 33268888
- DOI: https://doi.org/10.1038/s41586-020-2990-5
- Primary Citation of Related Structures:
6Z47 - PubMed Abstract:
Myosin-2 is essential for processes as diverse as cell division and muscle contraction. Dephosphorylation of its regulatory light chain promotes an inactive, 'shutdown' state with the filament-forming tail folded onto the two heads 1 , which prevents filament formation and inactivates the motors 2 . The mechanism by which this happens is unclear. Here we report a cryo-electron microscopy structure of shutdown smooth muscle myosin with a resolution of 6 Å in the head region. A pseudo-atomic model, obtained by flexible fitting of crystal structures into the density and molecular dynamics simulations, describes interaction interfaces at the atomic level. The N-terminal extension of one regulatory light chain interacts with the tail, and the other with the partner head, revealing how the regulatory light chains stabilize the shutdown state in different ways and how their phosphorylation would allow myosin activation. Additional interactions between the three segments of the coiled coil, the motor domains and the light chains stabilize the shutdown molecule. The structure of the lever in each head is competent to generate force upon activation. This shutdown structure is relevant to all isoforms of myosin-2 and provides a framework for understanding their disease-causing mutations.
- The Astbury Centre for Structural and Molecular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, UK.
Organizational Affiliation: