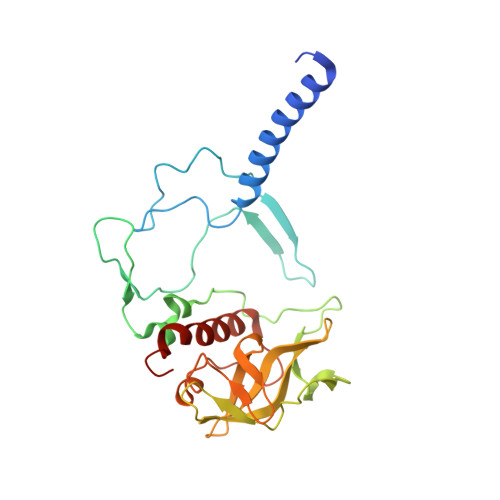
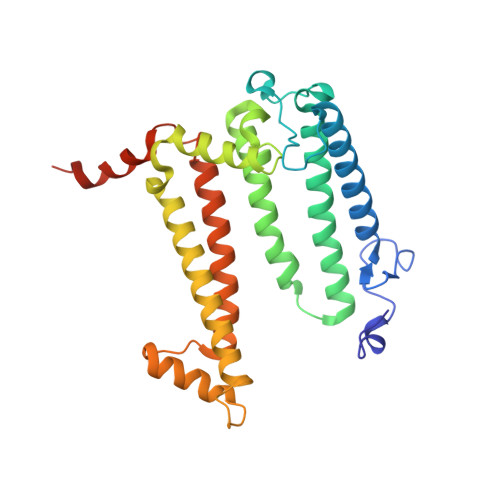
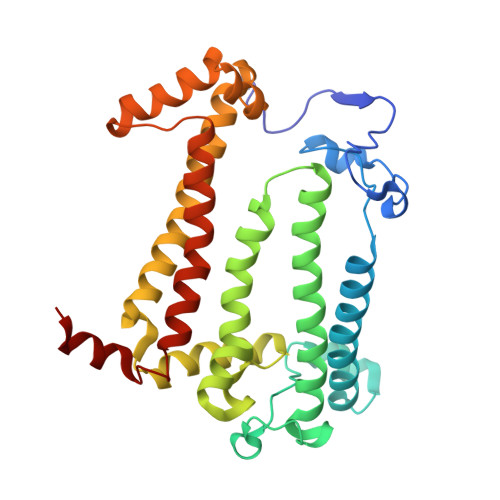
Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Selikhanov, G., Fufina, T., Vasilieva, L., Betzel, C., Gabdulkhakov, A.(2020) IUCrJ 7: 1084-1091
- PubMed: 33209319
- DOI: https://doi.org/10.1107/S2052252520012142
- Primary Citation of Related Structures:
6Z02, 6Z1J, 6Z27 - PubMed Abstract:
With the recent developments in the field of free-electron-laser-based serial femtosecond crystallography, the necessity to obtain a large number of high-quality crystals has emerged. In this work crystallization techniques were selected, tested and optimized for the lipid mesophase crystallization of the Rhodobacter sphaeroides membrane pigment-protein complex, known as the photosynthetic reaction center (RC). Novel approaches for lipid sponge phase crystallization in comparatively large volumes using Hamilton gas-tight glass syringes and plastic pipetting tips are described. An analysis of RC crystal structures obtained by lipid mesophase crystallization revealed non-native ligands that displaced the native electron-transfer cofactors (carotenoid sphero-idene and a ubi-quinone molecule) from their binding pockets. These ligands were identified and were found to be lipids that are major mesophase components. The selection of distinct co-crystallization conditions with the missing cofactors facilitated the restoration of sphero-idene in its binding site.
- Institute of Protein Research, Russian Academy of Sciences, Institutskaya 4, Puschino, Moscow region 142290, Russian Federation.
Organizational Affiliation: