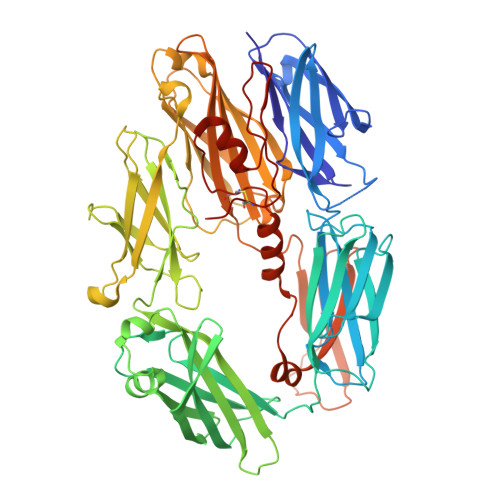
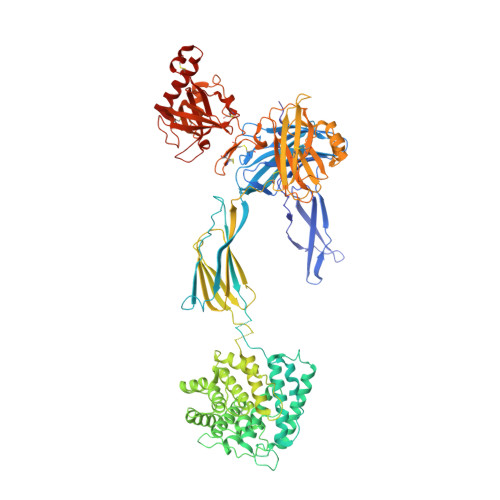
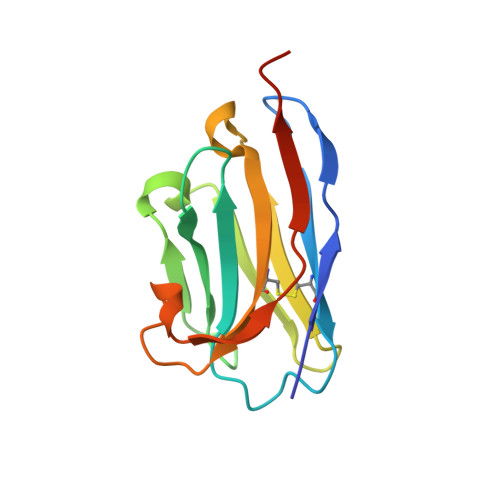
Complement Receptor 3 Forms a Compact High-Affinity Complex with iC3b.
Jensen, R.K., Bajic, G., Sen, M., Springer, T.A., Vorup-Jensen, T., Andersen, G.R.(2021) J Immunol 206: 3032-3042
- PubMed: 34117107
- DOI: https://doi.org/10.4049/jimmunol.2001208
- Primary Citation of Related Structures:
6YO6 - PubMed Abstract:
Complement receptor 3 (CR3, also known as Mac-1, integrin α M β 2 , or CD11b/CD18) is expressed on a subset of myeloid and certain activated lymphoid cells. CR3 is essential for the phagocytosis of complement-opsonized particles such as pathogens and apoptotic or necrotic cells opsonized with the complement fragment iC3b and, to a lesser extent, C3dg. Although the interaction between the iC3b thioester domain and the ligand binding CR3 α M I-domain is structurally and functionally well characterized, the nature of additional CR3-iC3b interactions required for phagocytosis of complement-opsonized objects remains obscure. In this study, we analyzed the interaction between iC3b and the 150-kDa headpiece fragment of the CR3 ectodomain. Surface plasmon resonance experiments demonstrated a 30 nM affinity of the CR3 headpiece for iC3b compared with 515 nM for the iC3b thioester domain, whereas experiments monitoring binding of iC3b to CR3-expressing cells suggested an affinity of 50 nM for the CR3-iC3b interaction. Small angle x-ray scattering analysis revealed that iC3b adopts an extended but preferred conformation in solution. Upon interaction with CR3, iC3b rearranges to form a compact receptor-ligand complex. Overall, the data suggest that the iC3b-CR3 interaction is of high affinity and relies on minor contacts formed between CR3 and regions outside the iC3b thioester domain. Our results rationalize the more efficient phagocytosis elicited by iC3b than by C3dg and pave the way for the development of specific therapeutics for the treatment of inflammatory and neurodegenerative diseases that do not interfere with the recognition of noncomplement CR3 ligands.
- Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark.
Organizational Affiliation: