Ancient and conserved functional interplay between Bcl-2 family proteins in the mitochondrial pathway of apoptosis.
Popgeorgiev, N., Sa, J.D., Jabbour, L., Banjara, S., Nguyen, T.T.M., Akhavan-E-Sabet, A., Gadet, R., Ralchev, N., Manon, S., Hinds, M.G., Osigus, H.J., Schierwater, B., Humbert, P.O., Rimokh, R., Gillet, G., Kvansakul, M.(2020) Sci Adv 6
- PubMed: 32998881
- DOI: https://doi.org/10.1126/sciadv.abc4149
- Primary Citation of Related Structures:
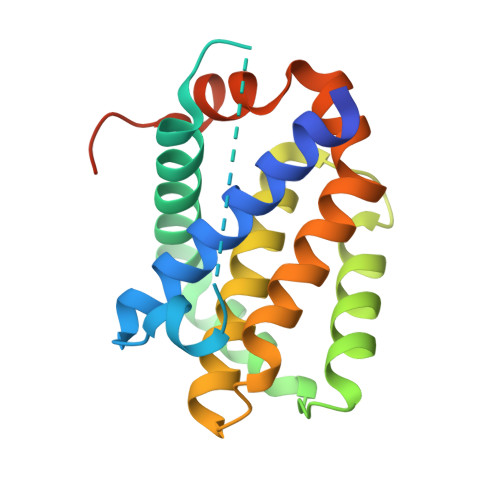
6YLD, 6YLI - PubMed Abstract:
In metazoans, Bcl-2 family proteins are major regulators of mitochondrially mediated apoptosis; however, their evolution remains poorly understood. Here, we describe the molecular characterization of the four members of the Bcl-2 family in the most primitive metazoan, Trichoplax adhaerens All four trBcl-2 homologs are multimotif Bcl-2 group, with trBcl-2L1 and trBcl-2L2 being highly divergent antiapoptotic Bcl-2 members, whereas trBcl-2L3 and trBcl-2L4 are homologs of proapoptotic Bax and Bak, respectively. trBax expression permeabilizes the mitochondrial outer membrane, while trBak operates as a BH3-only sensitizer repressing antiapoptotic activities of trBcl-2L1 and trBcl-2L2. The crystal structure of a trBcl-2L2:trBak BH3 complex reveals that trBcl-2L2 uses the canonical Bcl-2 ligand binding groove to sequester trBak BH3, indicating that the structural basis for apoptosis control is conserved from T. adhaerens to mammals. Finally, we demonstrate that both trBax and trBak BH3 peptides bind selectively to human Bcl-2 homologs to sensitize cancer cells to chemotherapy treatment.
- Université de Lyon, Centre de recherche en cancérologie de Lyon, U1052 INSERM, UMR CNRS 5286, Université Lyon I, Centre Léon Bérard, 28 rue Laennec, 69008 Lyon, France. nikolay.popgeorgiev@univ-lyon1.fr germain.gillet@univ-lyon1.fr m.kvansakul@latrobe.edu.au.
Organizational Affiliation: