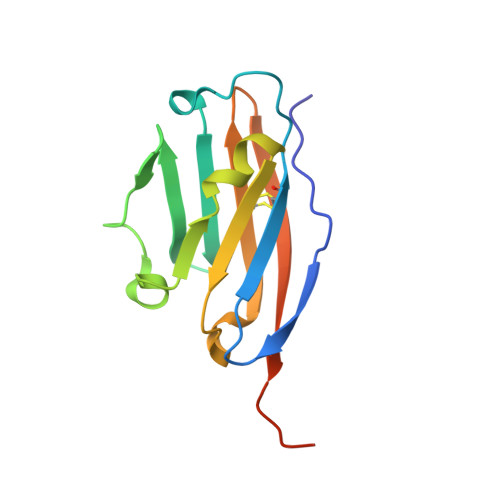
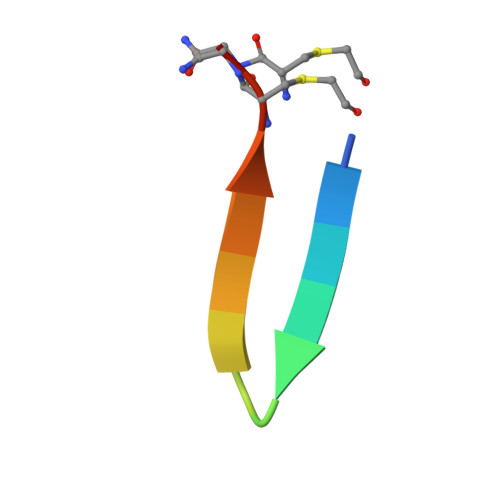
Macrocyclic Peptide Inhibitor of PD-1/PD-L1 Immune Checkpoint
Magiera-Mularz, K., Kuska, K., Skalniak, L., Grudnik, P., Musielak, B., Plewka, J., Kocik, J., Stec, M., Weglarczyk, K., Sala, D., Wladyka, B., Siedlar, M., Holak, T.A., Dubin, G.(2020) Adv Ther