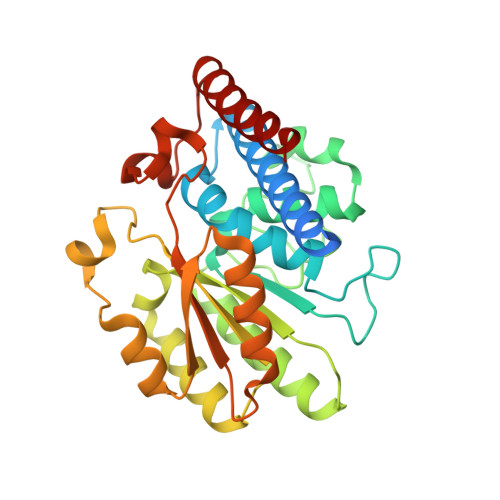
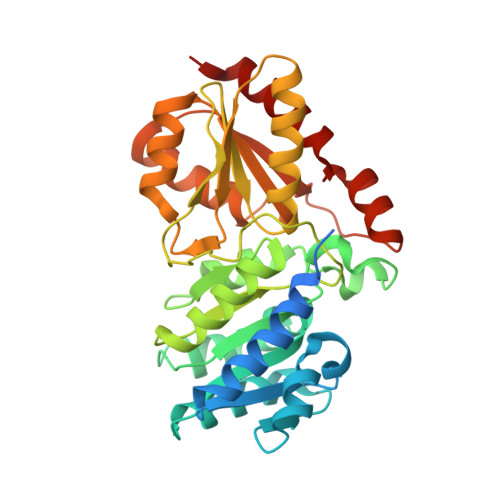
A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans : Structure and Biochemical Characterization.
James, P., Isupov, M.N., De Rose, S.A., Sayer, C., Cole, I.S., Littlechild, J.A.(2020) Front Microbiol 11: 592353-592353
- PubMed: 33193259
- DOI: https://doi.org/10.3389/fmicb.2020.592353
- Primary Citation of Related Structures:
6YAJ, 6YAK - PubMed Abstract:
A novel transketolase has been reconstituted from two separate polypeptide chains encoded by a 'split-gene' identified in the genome of the hyperthermophilic bacterium, Carboxydothermus hydrogenoformans . The reconstituted active α 2 β 2 tetrameric enzyme has been biochemically characterized and its activity has been determined using a range of aldehydes including glycolaldehyde, phenylacetaldehyde and cyclohexanecarboxaldehyde as the ketol acceptor and hydroxypyruvate as the donor. This reaction proceeds to near 100% completion due to the release of the product carbon dioxide and can be used for the synthesis of a range of sugars of interest to the pharmaceutical industry. This novel reconstituted transketolase is thermally stable with no loss of activity after incubation for 1 h at 70°C and is stable after 1 h incubation with 50% of the organic solvents methanol, ethanol, isopropanol, DMSO, acetonitrile and acetone. The X-ray structure of the holo reconstituted α 2 β 2 tetrameric transketolase has been determined to 1.4 Å resolution. In addition, the structure of an inactive tetrameric β 4 protein has been determined to 1.9 Å resolution. The structure of the active reconstituted α 2 β 2 enzyme has been compared to the structures of related enzymes; the E1 component of the pyruvate dehydrogenase complex and D-xylulose-5-phosphate synthase, in an attempt to rationalize differences in structure and substrate specificity between these enzymes. This is the first example of a reconstituted 'split-gene' transketolase to be biochemically and structurally characterized allowing its potential for industrial biocatalysis to be evaluated.
- Henry Wellcome Building for Biocatalysis, Biosciences, University of Exeter, Exeter, United Kingdom.
Organizational Affiliation: