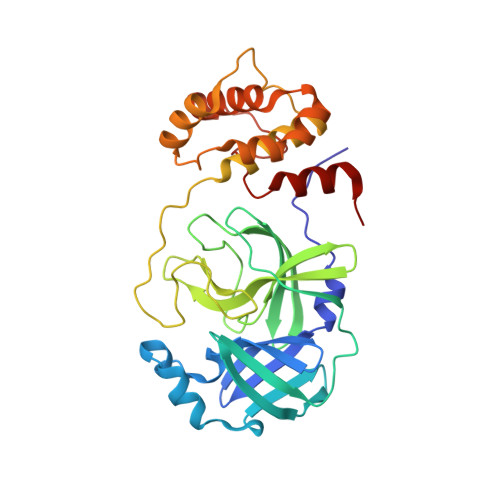
COVID-19 main protease with unliganded active site
Owen, C.D., Lukacik, P., Strain-Damerell, C.M., Douangamath, A., Powell, A.J., Fearon, D., Brandao-Neto, J., Crawshaw, A.D., Aragao, D., Williams, M., Flaig, R., Hall, D., McAauley, K., Stuart, D.I., von Delft, F., Walsh, M.A.To be published.