Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
Langston, S.P., Grossman, S., England, D., Afroze, R., Bence, N., Bowman, D., Bump, N., Chau, R., Chuang, B.C., Claiborne, C., Cohen, L., Connolly, K., Duffey, M., Durvasula, N., Freeze, S., Gallery, M., Galvin, K., Gaulin, J., Gershman, R., Greenspan, P., Grieves, J., Guo, J., Gulavita, N., Hailu, S., He, X., Hoar, K., Hu, Y., Hu, Z., Ito, M., Kim, M.S., Lane, S.W., Lok, D., Lublinsky, A., Mallender, W., McIntyre, C., Minissale, J., Mizutani, H., Mizutani, M., Molchinova, N., Ono, K., Patil, A., Qian, M., Riceberg, J., Shindi, V., Sintchak, M.D., Song, K., Soucy, T., Wang, Y., Xu, H., Yang, X., Zawadzka, A., Zhang, J., Pulukuri, S.M.(2021) J Med Chem 64: 2501-2520
- PubMed: 33631934
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01491
- Primary Citation of Related Structures:
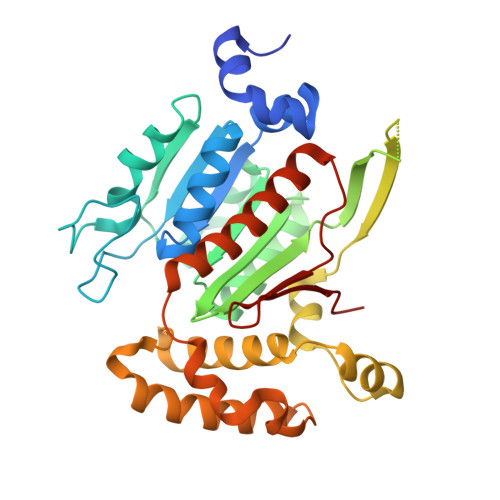
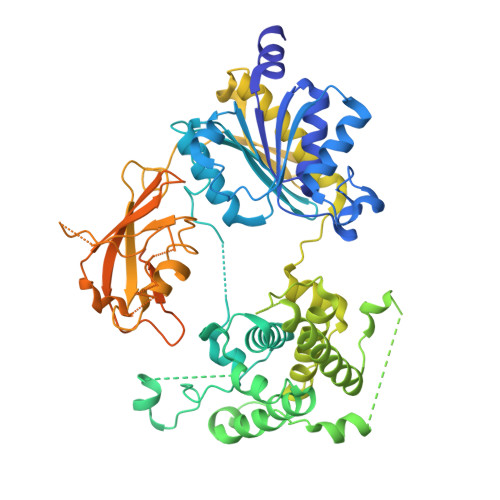
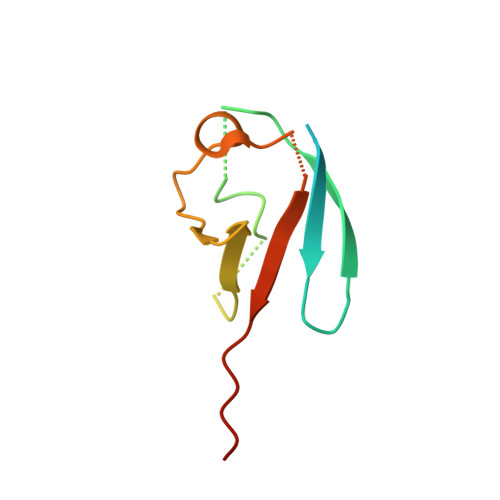
6XOG, 6XOH, 6XOI - PubMed Abstract:
SUMOylation is a reversible post-translational modification that regulates protein function through covalent attachment of small ubiquitin-like modifier (SUMO) proteins. The process of SUMOylating proteins involves an enzymatic cascade, the first step of which entails the activation of a SUMO protein through an ATP-dependent process catalyzed by SUMO-activating enzyme (SAE). Here, we describe the identification of TAK-981, a mechanism-based inhibitor of SAE which forms a SUMO-TAK-981 adduct as the inhibitory species within the enzyme catalytic site. Optimization of selectivity against related enzymes as well as enhancement of mean residence time of the adduct were critical to the identification of compounds with potent cellular pathway inhibition and ultimately a prolonged pharmacodynamic effect and efficacy in preclinical tumor models, culminating in the identification of the clinical molecule TAK-981.
- Millennium Pharmaceuticals, a wholly owned subsidiary of Takeda Pharmaceuticals Company Ltd., Cambridge, Massachusetts 02139, United States.
Organizational Affiliation: