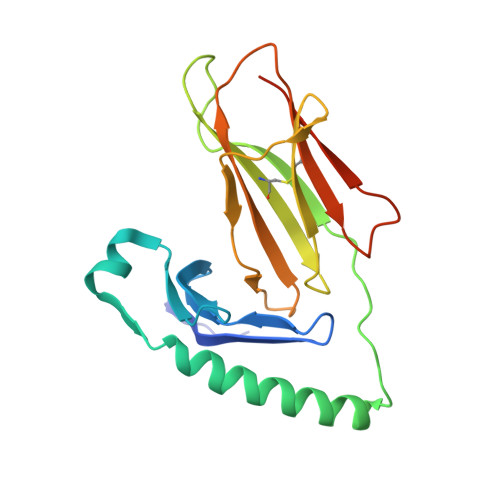
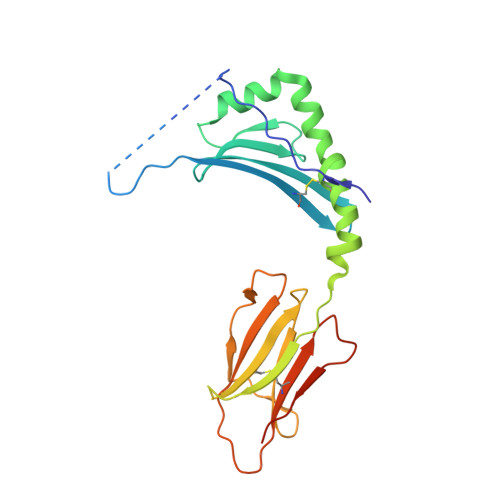
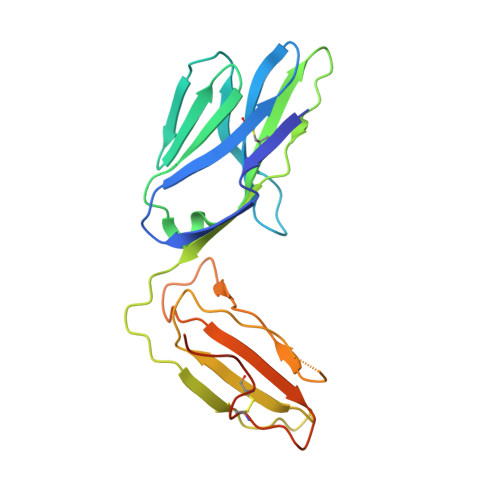
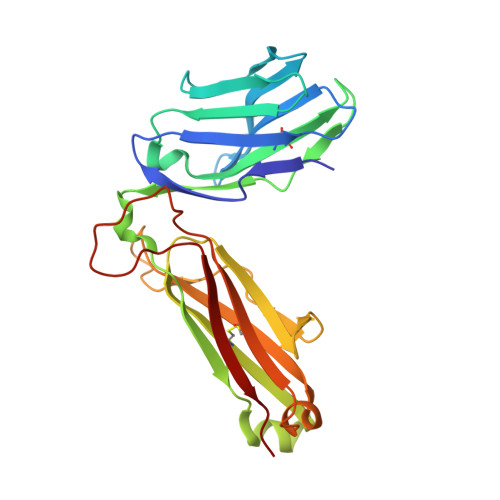
T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Tran, M.T., Faridi, P., Lim, J.J., Ting, Y.T., Onwukwe, G., Bhattacharjee, P., Jones, C.M., Tresoldi, E., Cameron, F.J., La Gruta, N.L., Purcell, A.W., Mannering, S.I., Rossjohn, J., Reid, H.H.(2021) Nat Commun 12: 5110-5110
- PubMed: 34433824
- DOI: https://doi.org/10.1038/s41467-021-25404-x
- Primary Citation of Related Structures:
6XC9, 6XCO, 6XCP - PubMed Abstract:
HLA-DQ8, a genetic risk factor in type I diabetes (T1D), presents hybrid insulin peptides (HIPs) to autoreactive CD4+ T cells. The abundance of spliced peptides binding to HLA-DQ8 and how they are subsequently recognised by the autoreactive T cell repertoire is unknown. Here we report, the HIP (GQVELGGGNAVEVLK), derived from splicing of insulin and islet amyloid polypeptides, generates a preferred peptide-binding motif for HLA-DQ8. HLA-DQ8-HIP tetramer + T cells from the peripheral blood of a T1D patient are characterised by repeated TRBV5 usage, which matches the TCR bias of CD4+ T cells reactive to the HIP peptide isolated from the pancreatic islets of a patient with T1D. The crystal structure of three TRBV5+ TCR-HLA-DQ8-HIP complexes shows that the TRBV5-encoded TCR β-chain forms a common landing pad on the HLA-DQ8 molecule. The N- and C-termini of the HIP is recognised predominantly by the TCR α-chain and TCR β-chain, respectively, in all three TCR ternary complexes. Accordingly, TRBV5 + TCR recognition of HIP peptides might occur via a 'polarised' mechanism, whereby each chain within the αβTCR heterodimer recognises distinct origins of the spliced peptide presented by HLA-DQ8.
- Infection and Immunity Program & Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, VIC, Australia.
Organizational Affiliation: