X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
Banerjee, S., Mahmud, F., Deng, S., Ma, L., Yun, M.K., Fakayode, S.O., Arnst, K.E., Yang, L., Chen, H., Wu, Z., Lukka, P.B., Parmar, K., Meibohm, B., White, S.W., Wang, Y., Li, W., Miller, D.D.(2021) J Med Chem 64: 13072-13095
- PubMed: 34406768
- DOI: https://doi.org/10.1021/acs.jmedchem.1c01202
- Primary Citation of Related Structures:
6X1C, 6X1E, 6X1F, 7LZ7, 7LZ8 - PubMed Abstract:
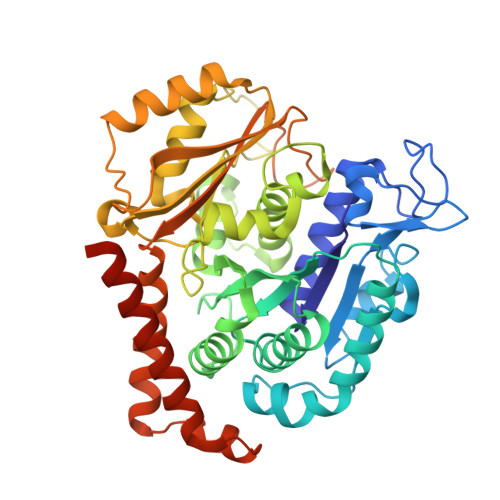
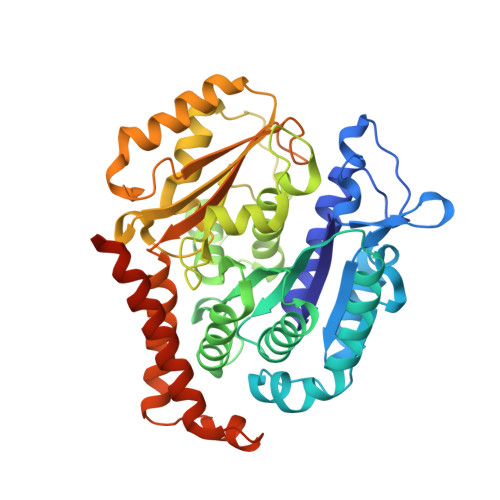
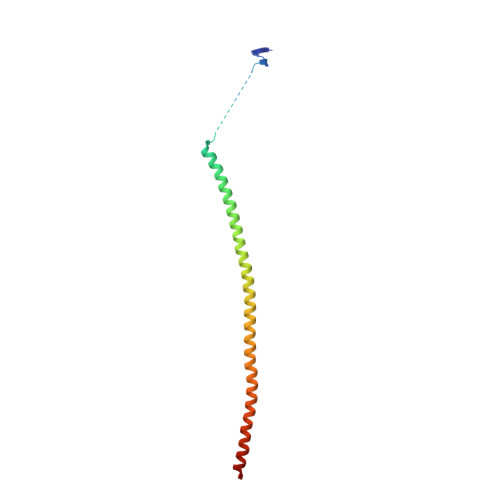
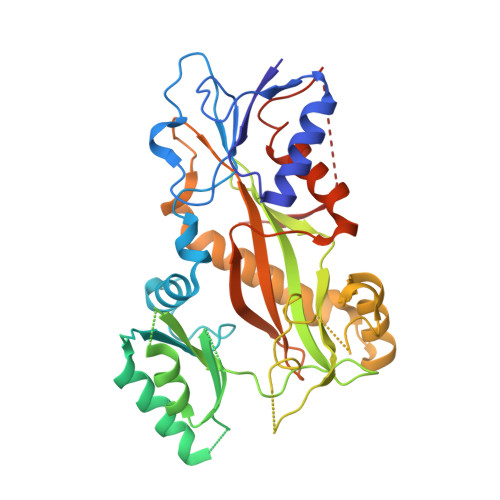
Small molecules that interact with the colchicine binding site in tubulin have demonstrated therapeutic efficacy in treating cancers. We report the design, syntheses, and antitumor efficacies of new analogues of pyridopyrimidine and hydroquinoxalinone compounds with improved drug-like characteristics. Eight analogues, 5j , 5k , 5l , 5m , 5n , 5r , 5t , and 5u , showed significant improvement in metabolic stability and demonstrated strong antiproliferative potency in a panel of human cancer cell lines, including melanoma, lung cancer, and breast cancer. We report crystal structures of tubulin in complex with five representative compounds, 5j , 5k , 5l , 5m , and 5t , providing direct confirmation for their binding to the colchicine site in tubulin. A quantitative structure-activity relationship analysis of the synthesized analogues showed strong ability to predict potency. In vivo , 5m (4 mg/kg) and 5t (5 mg/kg) significantly inhibited tumor growth as well as melanoma spontaneous metastasis into the lung and liver against a highly paclitaxel-resistant A375/TxR xenograft model.
- Department of Pharmaceutical Sciences, College of Pharmacy, University of Tennessee Health Science Center, Memphis, Tennessee 38163, United States.
Organizational Affiliation: