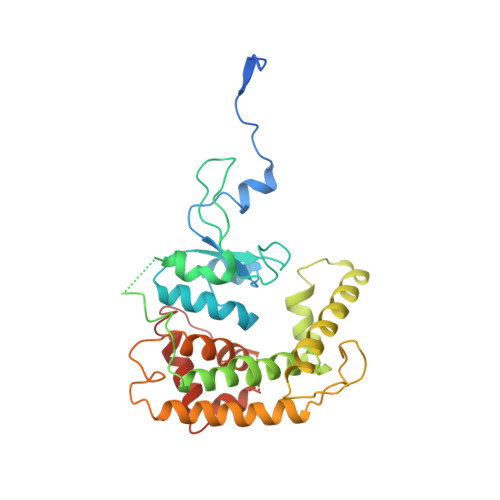
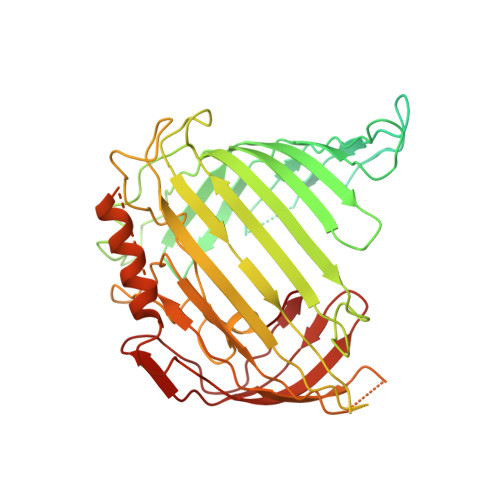
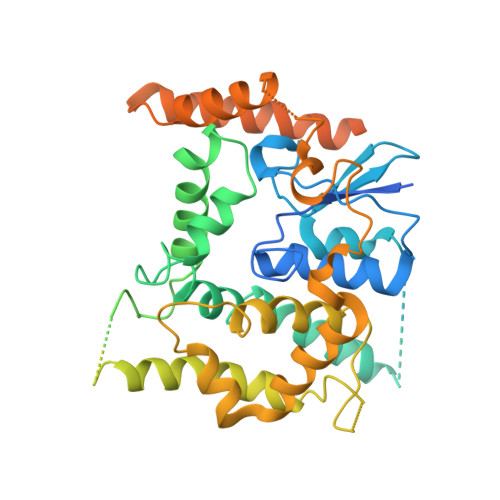
Structural insight into mitochondrial beta-barrel outer membrane protein biogenesis.
Diederichs, K.A., Ni, X., Rollauer, S.E., Botos, I., Tan, X., King, M.S., Kunji, E.R.S., Jiang, J., Buchanan, S.K.(2020) Nat Commun 11: 3290-3290
- PubMed: 32620929
- DOI: https://doi.org/10.1038/s41467-020-17144-1
- Primary Citation of Related Structures:
6WUH, 6WUJ, 6WUL, 6WUM, 6WUN, 6WUT - PubMed Abstract:
In mitochondria, β-barrel outer membrane proteins mediate protein import, metabolite transport, lipid transport, and biogenesis. The Sorting and Assembly Machinery (SAM) complex consists of three proteins that assemble as a 1:1:1 complex to fold β-barrel proteins and insert them into the mitochondrial outer membrane. We report cryoEM structures of the SAM complex from Myceliophthora thermophila, which show that Sam50 forms a 16-stranded transmembrane β-barrel with a single polypeptide-transport-associated (POTRA) domain extending into the intermembrane space. Sam35 and Sam37 are located on the cytosolic side of the outer membrane, with Sam35 capping Sam50, and Sam37 interacting extensively with Sam35. Sam35 and Sam37 each adopt a GST-like fold, with no functional, structural, or sequence similarity to their bacterial counterparts. Structural analysis shows how the Sam50 β-barrel opens a lateral gate to accommodate its substrates.
- Laboratory of Molecular Biology, National Institute of Diabetes & Digestive & Kidney Diseases, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD, 20892, USA.
Organizational Affiliation: