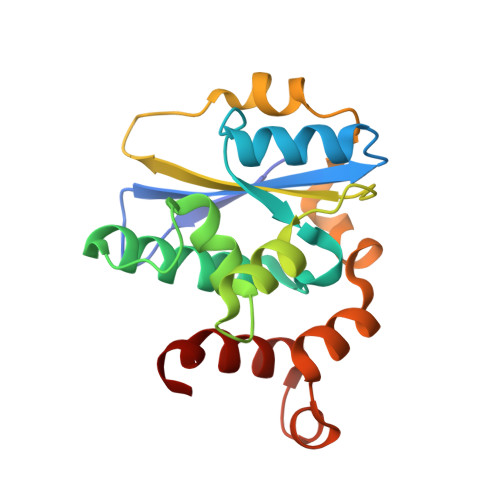
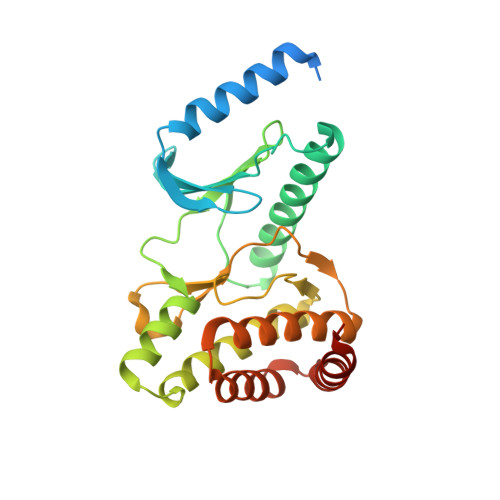
Crystal structure of the human PRPK-TPRKB complex.
Li, J., Ma, X., Banerjee, S., Chen, H., Ma, W., Bode, A.M., Dong, Z.(2021) Commun Biol 4: 167-167
- PubMed: 33547416
- DOI: https://doi.org/10.1038/s42003-021-01683-4
- Primary Citation of Related Structures:
6WQX - PubMed Abstract:
Mutations of the p53-related protein kinase (PRPK) and TP53RK-binding protein (TPRKB) cause Galloway-Mowat syndrome (GAMOS) and are found in various human cancers. We have previously shown that small compounds targeting PRPK showed anti-cancer activity against colon and skin cancer. Here we present the 2.53 Å crystal structure of the human PRPK-TPRKB-AMPPNP (adenylyl-imidodiphosphate) complex. The structure reveals details in PRPK-AMPPNP coordination and PRPK-TPRKB interaction. PRPK appears in an active conformation, albeit lacking the conventional kinase activation loop. We constructed a structural model of the human EKC/KEOPS complex, composed of PRPK, TPRKB, OSGEP, LAGE3, and GON7. Disease mutations in PRPK and TPRKB are mapped into the structure, and we show that one mutation, PRPK K238Nfs*2, lost the binding to OSGEP. Our structure also makes the virtual screening possible and paves the way for more rational drug design.
- The Hormel Institute, University of Minnesota, Austin, MN, 55912, USA.
Organizational Affiliation: