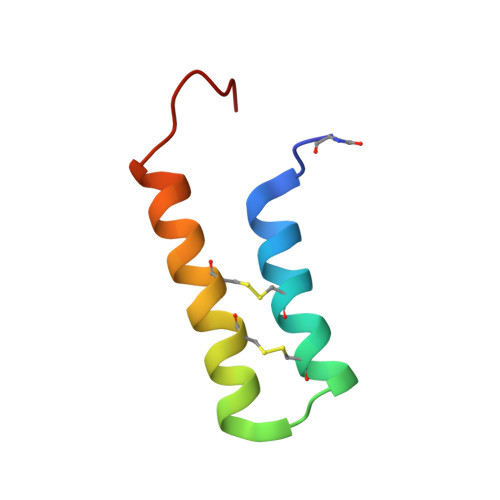
Defining the Familial Fold of the Vicilin-Buried Peptide Family.
Payne, C.D., Vadlamani, G., Fisher, M.F., Zhang, J., Clark, R.J., Mylne, J.S., Rosengren, K.J.(2020) J Nat Prod 83: 3030-3040
- PubMed: 32997497
- DOI: https://doi.org/10.1021/acs.jnatprod.0c00594
- Primary Citation of Related Structures:
6WQJ, 6WQL - PubMed Abstract:
Plants and their seeds have been shown to be a rich source of cystine-stabilized peptides. Recently a new family of plant seed peptides whose sequences are buried within precursors for seed storage vicilins was identified. Members of this Vicilin-Buried Peptide (VBP) family are found in distantly related plant species including the monocot date palm, as well as dicotyledonous species like pumpkin and sesame. Genetic evidence for their widespread occurrence indicates that they are of ancient origin. Limited structural studies have been conducted on VBP family members, but two members have been shown to adopt a helical hairpin fold. We present an extensive characterization of VBPs using solution NMR spectroscopy, to better understand their structural features. Four peptides were produced by solid phase peptide synthesis and shown to favor a helix-loop-helix hairpin fold, as a result of the I-IV/II-III ladderlike connectivity of their disulfide bonds. Interhelical interactions, including hydrophobic contacts and salt bridges, are critical for the fold stability and control the angle at which the antiparallel α-helices interface. Activities reported for VBPs include trypsin inhibitory activity and inhibition of ribosomal function; however, their diverse structural features despite a common fold suggest that additional bioactivities yet to be revealed are likely.
- School of Biomedical Sciences, The University of Queensland, Brisbane, Queensland 4072, Australia.
Organizational Affiliation: