RNA Binding by the KTS Splice Variants of Wilms' Tumor Suppressor Protein WT1.
Nishikawa, T., Wojciak, J.M., Dyson, H.J., Wright, P.E.(2020) Biochemistry 59: 3889-3901
- PubMed: 32955251
- DOI: https://doi.org/10.1021/acs.biochem.0c00602
- Primary Citation of Related Structures:
6WLH - PubMed Abstract:
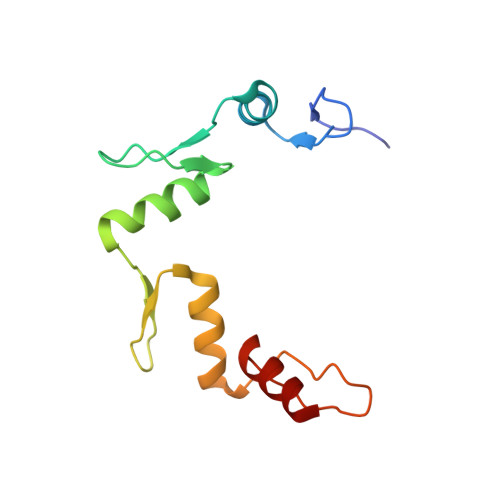
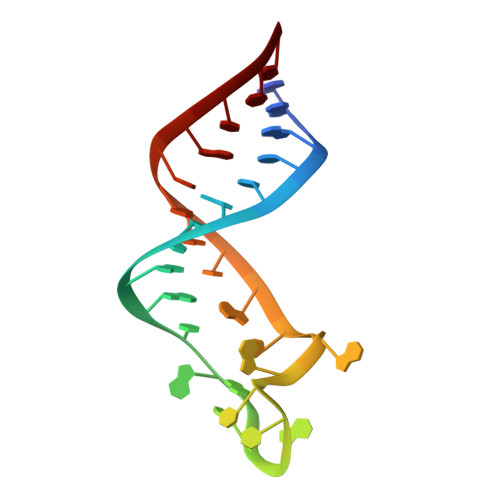
Wilms' tumor suppressor protein WT1 regulates the expression of multiple genes through binding of the Cys 2 -His 2 zinc finger domain to promoter sites. WT1 has also been proposed to be involved in post-transcriptional regulation, by binding to RNA using the same set of zinc fingers. WT1 has two major splice variants, where the Lys-Thr-Ser (KTS) tripeptide is inserted into the linker between the third and fourth zinc fingers. To obtain insights into the mechanism by which the different WT1 splice variants recognize both DNA and RNA, we have determined the solution structure of the WT1 (-KTS) zinc finger domain in complex with a 29mer stem-loop RNA. Zinc fingers 1-3 bind in a widened major groove favored by the presence of a bulge nucleotide in the double-stranded helical stem. Fingers 2 and 3 make specific contacts with the nucleobases in a conserved AUGG sequence in the helical stem. Nuclear magnetic resonance chemical shift mapping and relaxation analysis show that fingers 1-3 of the two splice variants (-KTS and +KTS) of WT1 form similar complexes with RNA. Finger 4 of the -KTS isoform interacts weakly with the RNA loop, an interaction that is abrogated in the +KTS isoform, and both isoforms bind with similar affinity to the RNA. In contrast, finger 4 is required for high-affinity binding to DNA and insertion of KTS into the linker of fingers 3 and 4 abrogates DNA binding. While finger 1 is required for RNA binding, it is dispensable for binding to consensus DNA sites.
- Department of Integrative Structural and Computational Biology and Skaggs Institute of Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States.
Organizational Affiliation: