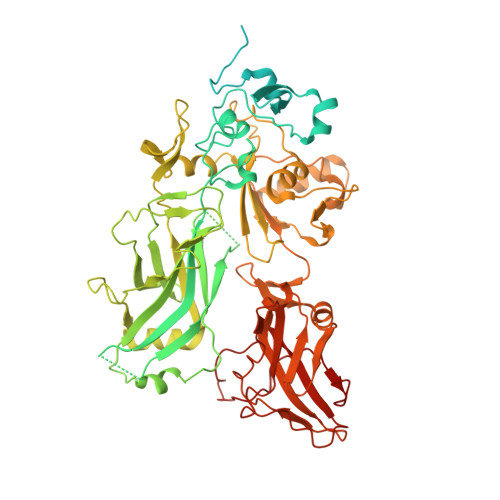
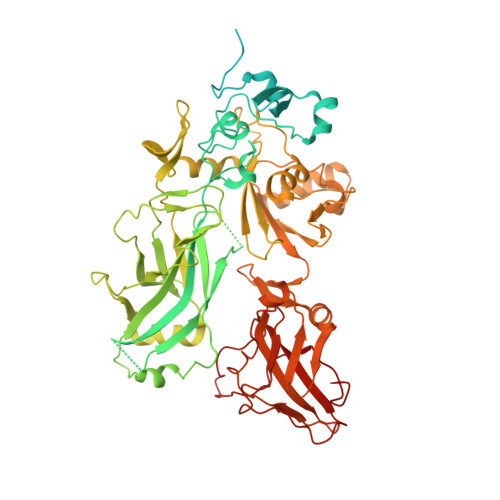
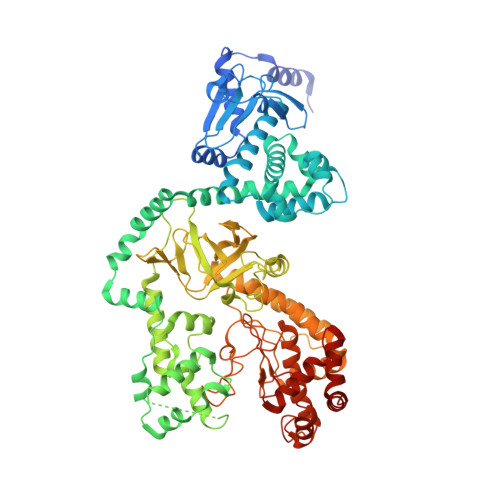
Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Zhou, K., Liu, S., Hardenbrook, N.J., Cui, Y., Krantz, B.A., Zhou, Z.H.(2020) Structure 28: 879-887.e3
- PubMed: 32521227
- DOI: https://doi.org/10.1016/j.str.2020.05.009
- Primary Citation of Related Structures:
6VRA, 6WJJ - PubMed Abstract:
Pathogenesis of anthrax disease involves two cytotoxic enzymes-edema factor (EF) and lethal factor (LF)-which are individually recruited by the protective antigen heptamer (PA 7 ) or octamer (PA 8 ) prechannel and subsequently translocated across channels formed on the endosomal membrane upon exposure to low pH. Here, we report the atomic structures of PA 8 prechannel-bound full-length EF and LF. In this pretranslocation state, the N-terminal segment of both factors refolds into an α helix engaged in the α clamp of the prechannel. Recruitment to the PA prechannel exposes an originally buried β strand of both toxins and enables domain organization of EF. Many interactions occur on domain interfaces in both PA prechannel-bound EF and LF, leading to toxin compaction prior to translocation. Our results provide key insights into the molecular mechanisms of translocation-coupled protein unfolding and translocation.
- California NanoSystems Institute, University of California, Los Angeles, CA 90095, USA.
Organizational Affiliation: