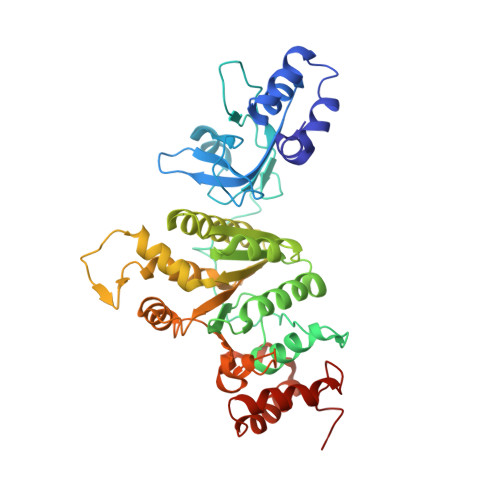
Crystal structure of the Escherichia coli transcription termination factor Rho.
Fan, C., Rees, D.C.(2020) Acta Crystallogr F Struct Biol Commun 76: 398-405
- PubMed: 32880587
- DOI: https://doi.org/10.1107/S2053230X20010572
- Primary Citation of Related Structures:
6WA8 - PubMed Abstract:
During the crystal structure analysis of an ATP-binding cassette (ABC) transporter overexpressed in Escherichia coli, a contaminant protein was crystallized. The identity of the contaminant was revealed by mass spectrometry to be the Escherichia coli transcription terminator factor Rho, structures of which had been previously determined in different conformational states. Although Rho was present at only ∼1% of the target protein (a bacterial homolog of the eukaryotic ABC transporter of mitochondria from Novosphingobium aromaticivorans; NaAtm1), it preferentially crystallized in space group C2 as thin plates that diffracted to 3.30 Å resolution. The structure of Rho in this crystal form exhibits a hexameric open-ring staircase conformation with bound ATP; this characteristic structure was also observed on electron-microscopy grids of the NaAtm1 preparation.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, 1200 East California Boulevard, Pasadena, CA 91125, USA.
Organizational Affiliation: