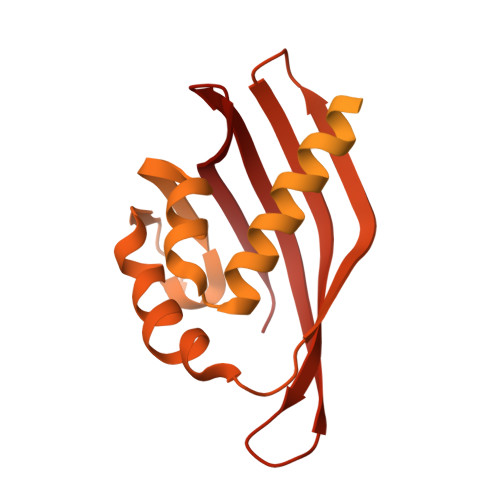
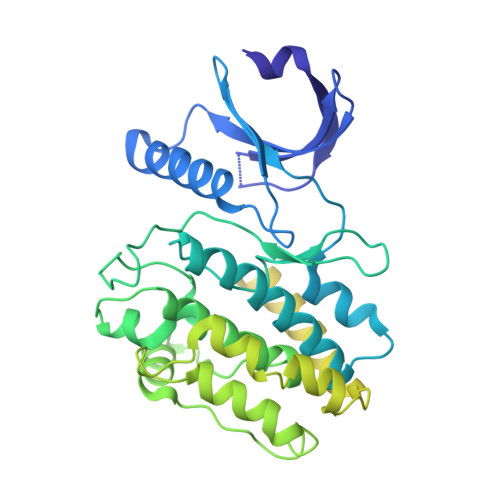
Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub-dependent regulation.
Sloutsky, R., Dziedzic, N., Dunn, M.J., Bates, R.M., Torres-Ocampo, A.P., Boopathy, S., Page, B., Weeks, J.G., Chao, L.H., Stratton, M.M.(2020) Sci Signal 13
- PubMed: 32694170
- DOI: https://doi.org/10.1126/scisignal.aaz0240
- Primary Citation of Related Structures:
6W4O, 6W4P - PubMed Abstract:
Calcium/calmodulin-dependent protein kinase II (CaMKII) plays a central role in Ca 2+ signaling throughout the body. In the hippocampus, CaMKII is required for learning and memory. Vertebrate genomes encode four CaMKII homologs: CaMKIIα, CaMKIIβ, CaMKIIγ, and CaMKIIδ. All CaMKIIs consist of a kinase domain, a regulatory segment, a variable linker region, and a hub domain, which is responsible for oligomerization. The four proteins differ primarily in linker length and composition because of extensive alternative splicing. Here, we report the heterogeneity of CaMKII transcripts in three complex samples of human hippocampus using deep sequencing. We showed that hippocampal cells contain a diverse collection of over 70 CaMKII transcripts from all four CaMKII-encoding genes. We characterized the Ca 2+ /CaM sensitivity of hippocampal CaMKII variants spanning a broad range of linker lengths and compositions. The effect of the variable linker on Ca 2+ /CaM sensitivity depended on the kinase and hub domains. Moreover, we revealed a previously uncharacterized role for the hub domain as an allosteric regulator of kinase activity, which may provide a pharmacological target for modulating CaMKII activity. Using small-angle x-ray scattering and single-particle cryo-electron microscopy (cryo-EM), we present evidence for extensive interactions between the kinase and the hub domains, even in the presence of a 30-residue linker. Together, these data suggest that Ca 2+ /CaM sensitivity in CaMKII is homolog dependent and includes substantial contributions from the hub domain. Our sequencing approach, combined with biochemistry, provides insights into understanding the complex pool of endogenous CaMKII splice variants.
- Department of Biochemistry and Molecular Biology, University of Massachusetts, Amherst, MA 01003, USA.
Organizational Affiliation: