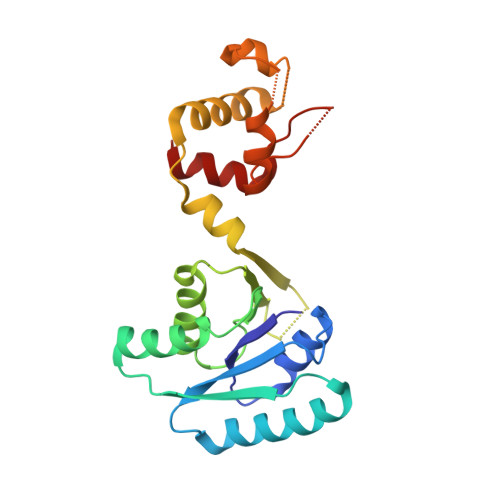
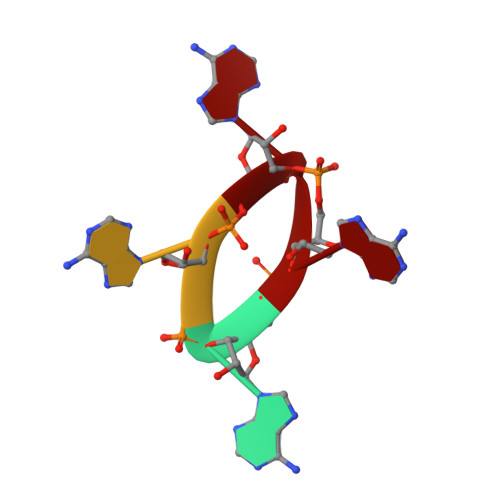
Cyclic Tetra-Adenylate (cA 4 ) Recognition by Csa3; Implications for an Integrated Class 1 CRISPR-Cas Immune Response in Saccharolobus solfataricus.
Charbonneau, A.A., Eckert, D.M., Gauvin, C.C., Lintner, N.G., Lawrence, C.M.(2021) Biomolecules 11
- PubMed: 34944496
- DOI: https://doi.org/10.3390/biom11121852
- Primary Citation of Related Structures:
6W11 - PubMed Abstract:
Csa3 family transcription factors are ancillary CRISPR-associated proteins composed of N-terminal CARF domains and C-terminal winged helix-turn-helix domains. The activity of Csa3 transcription factors is thought to be controlled by cyclic oligoadenyate (cOA) second messengers produced by type III CRISPR-Cas surveillance complexes. Here we show that Saccharolobus solfataricus Csa3a recognizes cyclic tetra-adenylate (cA 4 ) and that Csa3a lacks self-regulating "ring nuclease" activity present in some other CARF domain proteins. The crystal structure of the Csa3a/cA4 complex was also determined and the structural and thermodynamic basis for cA 4 recognition are described, as are conformational changes in Csa3a associated with cA 4 binding. We also characterized the effect of cA 4 on recognition of putative DNA binding sites. Csa3a binds to putative promoter sequences in a nonspecific, cooperative and cA 4 -independent manner, suggesting a more complex mode of transcriptional regulation. We conclude the Csa3a/cA 4 interaction represents a nexus between the type I and type III CRISPR-Cas systems present in S. solfataricus , and discuss the role of the Csa3/cA 4 interaction in coordinating different arms of this integrated class 1 immune system to mount a synergistic, highly orchestrated immune response.
- Department of Chemistry and Biochemistry, Montana State University, Bozeman, MT 59717, USA.
Organizational Affiliation: