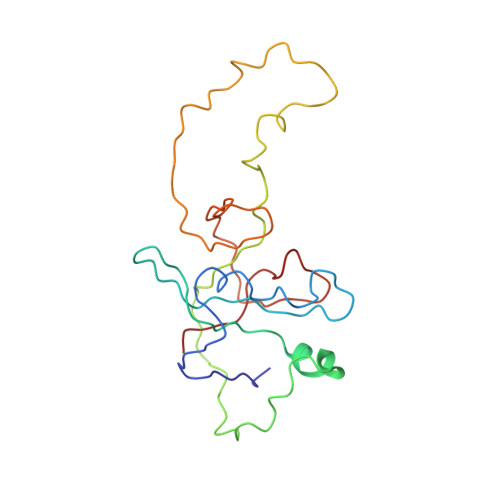
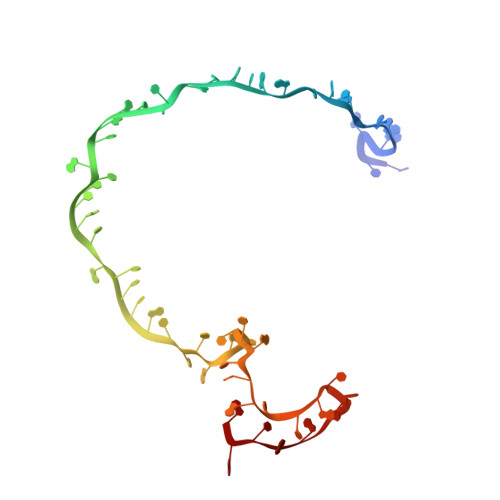
Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Zhang, K., Wang, S., Li, S., Zhu, Y., Pintilie, G.D., Mou, T.C., Schmid, M.F., Huang, Z., Chiu, W.(2020) Proc Natl Acad Sci U S A 117: 7176-7182
- PubMed: 32170016
- DOI: https://doi.org/10.1073/pnas.1922638117
- Primary Citation of Related Structures:
6VQV, 6VQW, 6VQX - PubMed Abstract:
Prokaryotes and viruses have fought a long battle against each other. Prokaryotes use CRISPR-Cas-mediated adaptive immunity, while conversely, viruses evolve multiple anti-CRISPR (Acr) proteins to defeat these CRISPR-Cas systems. The type I-F CRISPR-Cas system in Pseudomonas aeruginosa requires the crRNA-guided surveillance complex (Csy complex) to recognize the invading DNA. Although some Acr proteins against the Csy complex have been reported, other relevant Acr proteins still need studies to understand their mechanisms. Here, we obtain three structures of previously unresolved Acr proteins (AcrF9, AcrF8, and AcrF6) bound to the Csy complex using electron cryo-microscopy (cryo-EM), with resolution at 2.57 Å, 3.42 Å, and 3.15 Å, respectively. The 2.57-Å structure reveals fine details for each molecular component within the Csy complex as well as the direct and water-mediated interactions between proteins and CRISPR RNA (crRNA). Our structures also show unambiguously how these Acr proteins bind differently to the Csy complex. AcrF9 binds to key DNA-binding sites on the Csy spiral backbone. AcrF6 binds at the junction between Cas7.6f and Cas8f, which is critical for DNA duplex splitting. AcrF8 binds to a distinct position on the Csy spiral backbone and forms interactions with crRNA, which has not been seen in other Acr proteins against the Csy complex. Our structure-guided mutagenesis and biochemistry experiments further support the anti-CRISPR mechanisms of these Acr proteins. Our findings support the convergent consequence of inhibiting degradation of invading DNA by these Acr proteins, albeit with different modes of interactions with the type I-F CRISPR-Cas system.
- Department of Bioengineering, James H. Clark Center, Stanford University, Stanford, CA 94305.
Organizational Affiliation: