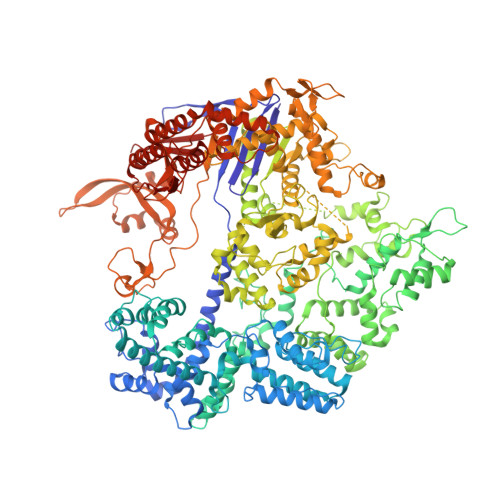
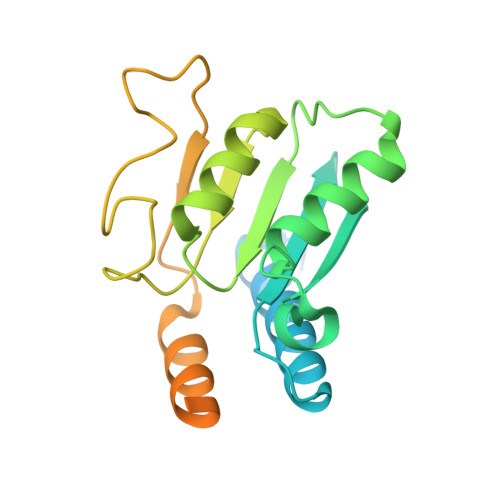
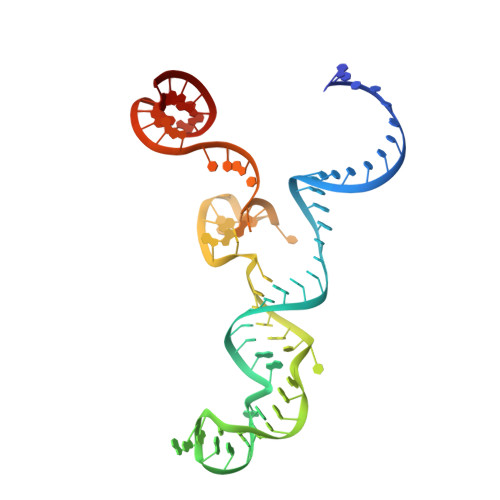
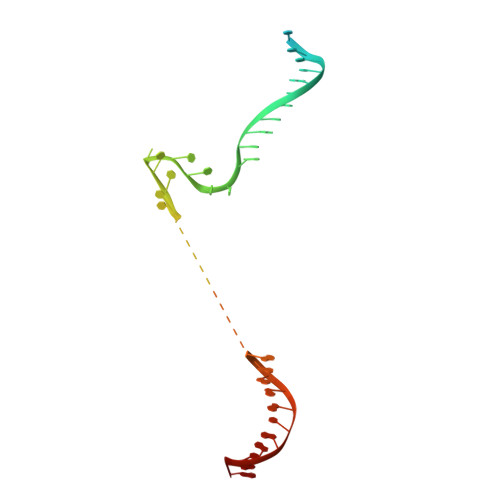
DNA capture by a CRISPR-Cas9-guided adenine base editor.
Lapinaite, A., Knott, G.J., Palumbo, C.M., Lin-Shiao, E., Richter, M.F., Zhao, K.T., Beal, P.A., Liu, D.R., Doudna, J.A.(2020) Science 369: 566-571
- PubMed: 32732424
- DOI: https://doi.org/10.1126/science.abb1390
- Primary Citation of Related Structures:
6VPC - PubMed Abstract:
CRISPR-Cas-guided base editors convert A•T to G•C, or C•G to T•A, in cellular DNA for precision genome editing. To understand the molecular basis for DNA adenosine deamination by adenine base editors (ABEs), we determined a 3.2-angstrom resolution cryo-electron microscopy structure of ABE8e in a substrate-bound state in which the deaminase domain engages DNA exposed within the CRISPR-Cas9 R-loop complex. Kinetic and structural data suggest that ABE8e catalyzes DNA deamination up to ~1100-fold faster than earlier ABEs because of mutations that stabilize DNA substrates in a constrained, transfer RNA-like conformation. Furthermore, ABE8e's accelerated DNA deamination suggests a previously unobserved transient DNA melting that may occur during double-stranded DNA surveillance by CRISPR-Cas9. These results explain ABE8e-mediated base-editing outcomes and inform the future design of base editors.
- Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Organizational Affiliation: