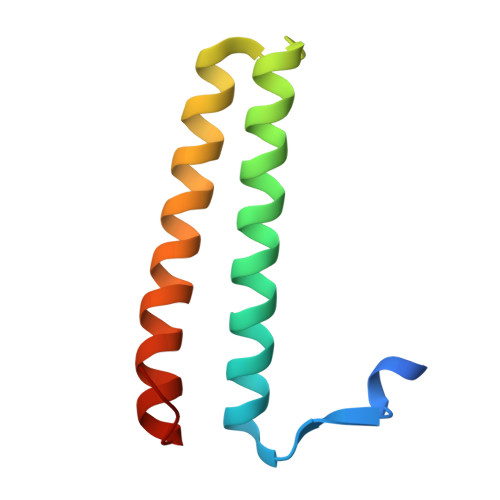
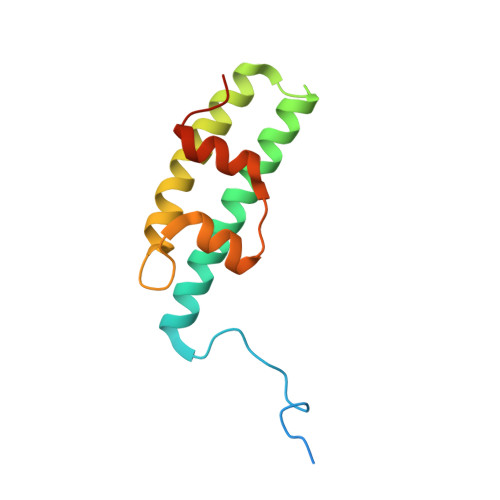
A helical assembly of human ESCRT-I scaffolds reverse-topology membrane scission.
Flower, T.G., Takahashi, Y., Hudait, A., Rose, K., Tjahjono, N., Pak, A.J., Yokom, A.L., Liang, X., Wang, H.G., Bouamr, F., Voth, G.A., Hurley, J.H.(2020) Nat Struct Mol Biol 27: 570-580
- PubMed: 32424346
- DOI: https://doi.org/10.1038/s41594-020-0426-4
- Primary Citation of Related Structures:
6VME - PubMed Abstract:
The ESCRT complexes drive membrane scission in HIV-1 release, autophagosome closure, multivesicular body biogenesis, cytokinesis, and other cell processes. ESCRT-I is the most upstream complex and bridges the system to HIV-1 Gag in virus release. The crystal structure of the headpiece of human ESCRT-I comprising TSG101-VPS28-VPS37B-MVB12A was determined, revealing an ESCRT-I helical assembly with a 12-molecule repeat. Electron microscopy confirmed that ESCRT-I subcomplexes form helical filaments in solution. Mutation of VPS28 helical interface residues blocks filament formation in vitro and autophagosome closure and HIV-1 release in human cells. Coarse-grained (CG) simulations of ESCRT assembly at HIV-1 budding sites suggest that formation of a 12-membered ring of ESCRT-I molecules is a geometry-dependent checkpoint during late stages of Gag assembly and HIV-1 budding and templates ESCRT-III assembly for membrane scission. These data show that ESCRT-I is not merely a bridging adaptor; it has an essential scaffolding and mechanical role in its own right.
- Department of Molecular and Cell Biology and California Institute for Quantitative Biosciences, University of California, Berkeley, Berkeley, CA, USA.
Organizational Affiliation: