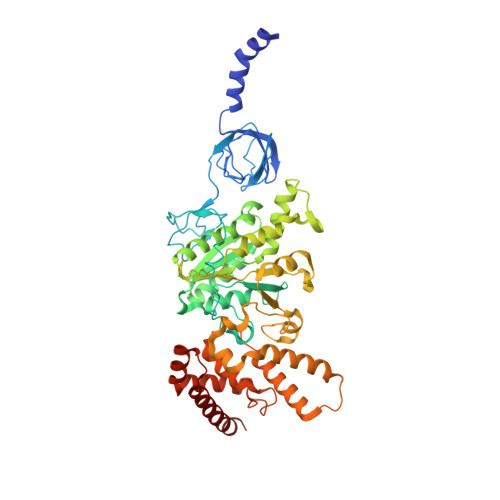
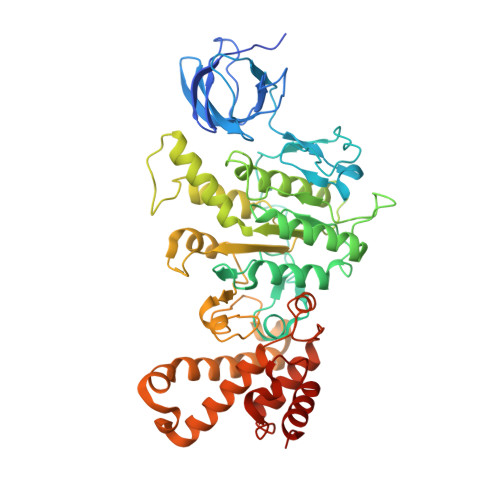
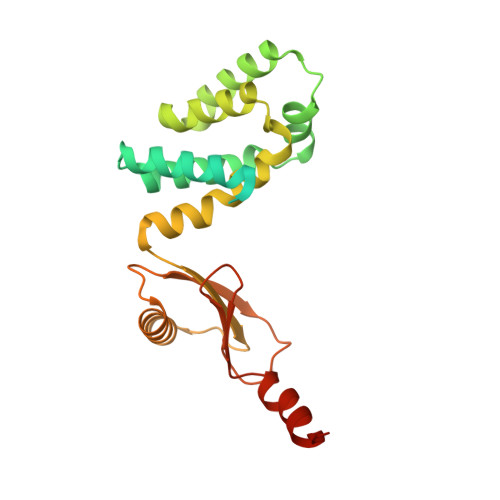
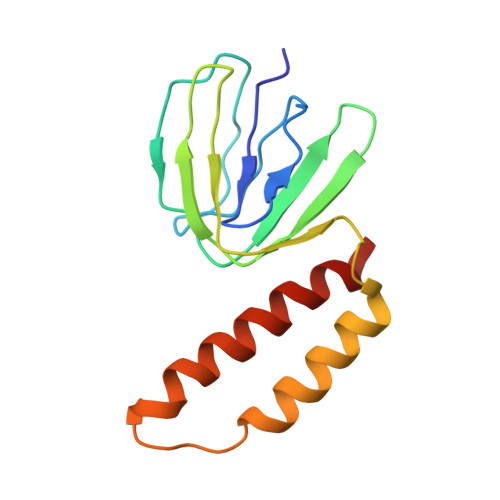
Structural basis of redox modulation on chloroplast ATP synthase.
Yang, J.H., Williams, D., Kandiah, E., Fromme, P., Chiu, P.L.(2020) Commun Biol 3: 482-482
- PubMed: 32879423
- DOI: https://doi.org/10.1038/s42003-020-01221-8
- Primary Citation of Related Structures:
6VM1, 6VM4, 6VMB, 6VMD, 6VMG, 6VOF, 6VOG, 6VOH, 6VOI, 6VOJ, 6VOK, 6VOL, 6VOM, 6VON, 6VOO - PubMed Abstract:
In higher plants, chloroplast ATP synthase has a unique redox switch on its γ subunit that modulates enzyme activity to limit ATP hydrolysis at night. To understand the molecular details of the redox modulation, we used single-particle cryo-EM to determine the structures of spinach chloroplast ATP synthase in both reduced and oxidized states. The disulfide linkage of the oxidized γ subunit introduces a torsional constraint to stabilize the two β hairpin structures. Once reduced, free cysteines alleviate this constraint, resulting in a concerted motion of the enzyme complex and a smooth transition between rotary states to facilitate the ATP synthesis. We added an uncompetitive inhibitor, tentoxin, in the reduced sample to limit the flexibility of the enzyme and obtained high-resolution details. Our cryo-EM structures provide mechanistic insight into the redox modulation of the energy regulation activity of chloroplast ATP synthase.
- Center for Applied Structural Discovery (CASD), Biodesign Institute, Arizona State University, Tempe, AZ, USA.
Organizational Affiliation: