Structural adaptation of oxygen tolerance in a key archaeal carbon fixation enzyme
Demirci, H., Wakatsuki, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Vinylacetyl-CoA Delta-isomerase | 502 | Nitrosopumilus maritimus SCM1 | Mutation(s): 0 Gene Names: Nmar_0207 EC: 5.3.3.3 | 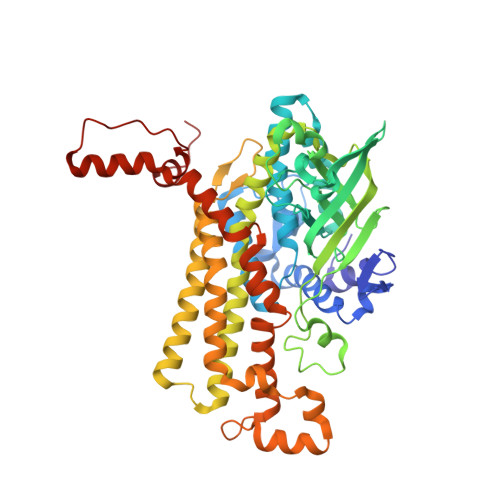 | |
UniProt | |||||
Find proteins for A9A1Y2 (Nitrosopumilus maritimus (strain SCM1)) Explore A9A1Y2 Go to UniProtKB: A9A1Y2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A9A1Y2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD (Subject of Investigation/LOI) Query on FAD | CA [auth D], F [auth A], N [auth B], V [auth C] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| SF4 Query on SF4 | BA [auth D], E [auth A], M [auth B], U [auth C] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | DA [auth D] G [auth A] H [auth A] O [auth B] T [auth C] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| FE Query on FE | EA [auth D], I [auth A], P [auth B], X [auth C] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| NA Query on NA | AA [auth C] GA [auth D] HA [auth D] K [auth A] L [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| UNL Query on UNL | FA [auth D], J [auth A], Q [auth B], Y [auth C] | Unknown ligand FKNQFGJONOIPTF-UHFFFAOYSA-N | |||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 87.53 | α = 90 |
| b = 72.954 | β = 98.38 |
| c = 180.517 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| PDB_EXTRACT | data extraction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, United States) | United States | 1231306 |