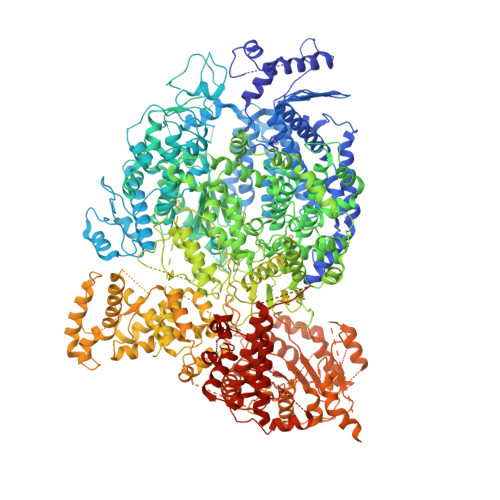
Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation.
Abdella, R., Aggarwal, M., Okura, T., Lamb, R.A., He, Y.(2020) Proc Natl Acad Sci U S A 117: 4931-4941
- PubMed: 32075920
- DOI: https://doi.org/10.1073/pnas.1919837117
- Primary Citation of Related Structures:
6V85, 6V86, 6VAG - PubMed Abstract:
Paramyxoviruses are enveloped, nonsegmented, negative-strand RNA viruses that cause a wide spectrum of human and animal diseases. The viral genome, packaged by the nucleoprotein (N), serves as a template for the polymerase complex, composed of the large protein (L) and the homo-tetrameric phosphoprotein (P). The ∼250-kDa L possesses all enzymatic activities necessary for its function but requires P in vivo. Structural information is available for individual P domains from different paramyxoviruses, but how P interacts with L and how that affects the activity of L is largely unknown due to the lack of high-resolution structures of this complex in this viral family. In this study we determined the structure of the L-P complex from parainfluenza virus 5 (PIV5) at 4.3-Å resolution using cryoelectron microscopy, as well as the oligomerization domain (OD) of P at 1.4-Å resolution using X-ray crystallography. P-OD associates with the RNA-dependent RNA polymerase domain of L and protrudes away from it, while the X domain of one chain of P is bound near the L nucleotide entry site. The methyltransferase (MTase) domain and the C-terminal domain (CTD) of L adopt a unique conformation, positioning the MTase active site immediately above the poly-ribonucleotidyltransferase domain and near the likely exit site for the product RNA 5' end. Our study reveals a potential mechanism that mononegavirus polymerases may employ to switch between transcription and genome replication. This knowledge will assist in the design and development of antivirals against paramyxoviruses.
- Department of Molecular Biosciences, Northwestern University, Evanston, IL 60208.
Organizational Affiliation: