A single-domain bispecific antibody targeting CD1d and the NKT T-cell receptor induces a potent antitumor response.
Shahine, A., Rossjohn, J.(2020) Nat Cancer
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Currently 6V80 does not have a validation slider image.
(2020) Nat Cancer
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Antigen-presenting glycoprotein CD1d | 347 | Homo sapiens | Mutation(s): 0 Gene Names: CD1D | 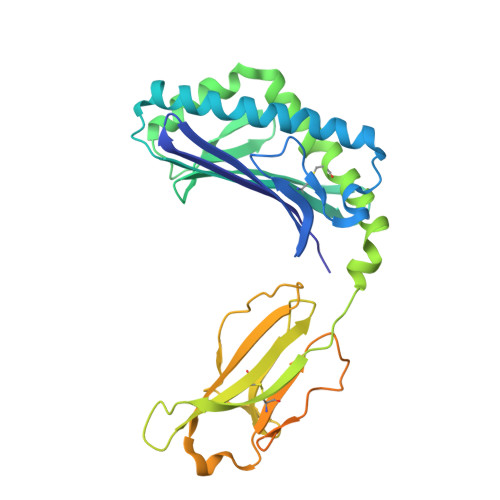 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P15813 (Homo sapiens) Explore P15813 Go to UniProtKB: P15813 | |||||
PHAROS: P15813 GTEx: ENSG00000158473 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P15813 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | Go to GlyGen: P15813-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | 100 | Homo sapiens | Mutation(s): 0 Gene Names: B2M, CDABP0092, HDCMA22P | 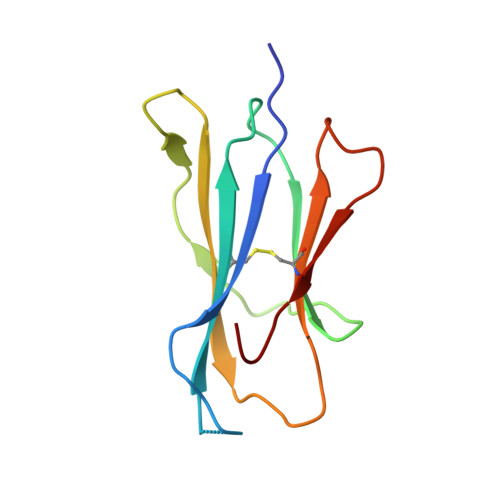 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61769 (Homo sapiens) Explore P61769 Go to UniProtKB: P61769 | |||||
PHAROS: P61769 GTEx: ENSG00000166710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61769 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| T cell receptor alpha variable 10, nkt tcr alpha chain fusion | 210 | Homo sapiens | Mutation(s): 0 Gene Names: TRAV10, B2M, HDCMA22P | 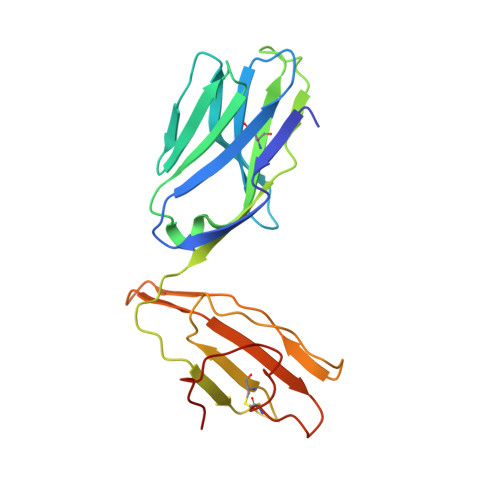 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for A0A0B4J240 (Homo sapiens) Explore A0A0B4J240 Go to UniProtKB: A0A0B4J240 | |||||
PHAROS: A0A0B4J240 GTEx: ENSG00000211784 | |||||
Find proteins for K7N5M3 (Homo sapiens) Explore K7N5M3 Go to UniProtKB: K7N5M3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | K7N5M3A0A0B4J240 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| nkt tcr beta chain | 241 | Homo sapiens | Mutation(s): 0 Gene Names: B2M, HDCMA22P | 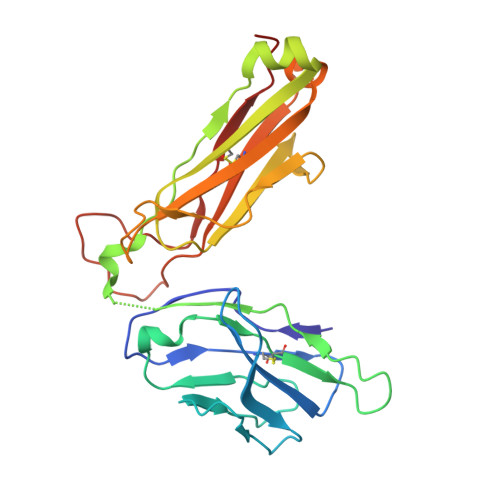 | |
UniProt | |||||
Find proteins for K7N5M4 (Homo sapiens) Explore K7N5M4 Go to UniProtKB: K7N5M4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | K7N5M4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody VHH ID12 | 119 | Lama glama | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AGH Query on AGH | M [auth A], N [auth F] | N-{(1S,2R,3S)-1-[(ALPHA-D-GALACTOPYRANOSYLOXY)METHYL]-2,3-DIHYDROXYHEPTADECYL}HEXACOSANAMIDE C50 H99 N O9 VQFKFAKEUMHBLV-BYSUZVQFSA-N |  | ||
| NAG Query on NAG | O [auth F], P [auth F] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 210.667 | α = 90 |
| b = 165.25 | β = 90 |
| c = 84.497 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
Currently 6V80 does not have a validation slider image.