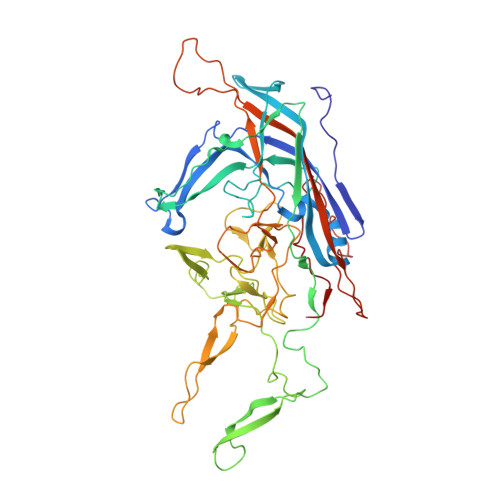
Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
Mietzsch, M., Barnes, C., Hull, J.A., Chipman, P., Xie, J., Bhattacharya, N., Sousa, D., McKenna, R., Gao, G., Agbandje-McKenna, M.(2020) J Virol 94
- PubMed: 31826994
- DOI: https://doi.org/10.1128/JVI.01769-19
- Primary Citation of Related Structures:
6O9R, 6V10, 6V12, 6V1G, 6V1T, 6V1Z - PubMed Abstract:
Adeno-associated viruses (AAVs) from clade E are often used as vectors in gene delivery applications. This clade includes rhesus isolate 10 (AAVrh.10) and 39 (AAVrh.39) which, unlike representative AAV8, are capable of crossing the blood-brain barrier (BBB), thereby enabling the delivery of therapeutic genes to the central nervous system. Here, the capsid structures of AAV8, AAVrh.10 and AAVrh.39 have been determined by cryo-electron microscopy and three-dimensional image reconstruction to 3.08-, 2.75-, and 3.39-Å resolution, respectively, to enable a direct structural comparison. AAVrh.10 and AAVrh.39 are 98% identical in amino acid sequence but only ∼93.5% identical to AAV8. However, the capsid structures of all three viruses are similar, with only minor differences observed in the previously described surface variable regions, suggesting that specific residues S269 and N472, absent in AAV8, may confer the ability to cross the BBB in AAVrh.10 and AAVrh.39. Head-to-head comparison of empty and genome-containing particles showed DNA ordered in the previously described nucleotide-binding pocket, supporting the suggested role of this pocket in DNA packaging for the Dependoparvovirus The structural characterization of these viruses provides a platform for future vector engineering efforts toward improved gene delivery success with respect to specific tissue targeting, transduction efficiency, antigenicity, or receptor retargeting. IMPORTANCE Recombinant adeno-associated virus vectors (rAAVs), based on AAV8 and AAVrh.10, have been utilized in multiple clinical trials to treat different monogenetic diseases. The closely related AAVrh.39 has also shown promise in vivo As recently attained for other AAV biologics, e.g., Luxturna and Zolgensma, based on AAV2 and AAV9, respectively, the vectors in this study will likely gain U.S. Food and Drug Administration approval for commercialization in the near future. This study characterized the capsid structures of these clinical vectors at atomic resolution using cryo-electron microscopy and image reconstruction for comparative analysis. The analysis suggested two key residues, S269 and N472, as determinants of BBB crossing for AAVrh.10 and AAVrh.39, a feature utilized for central nervous system delivery of therapeutic genes. The structure information thus provides a platform for engineering to improve receptor retargeting or tissue specificity. These are important challenges in the field that need attention. Capsid structure information also provides knowledge potentially applicable for regulatory product approval.
- Department of Biochemistry and Molecular Biology, Center for Structural Biology, McKnight Brain, Institute, College of Medicine, University of Florida, Gainesville, Florida, USA.
Organizational Affiliation: